Abstract
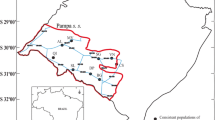
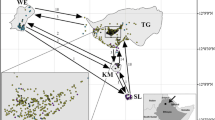
Within and among population gene flow is a central aspect of the evolutionary history of ecosystems and essential for the potential for adaptive evolution of populations. We employed nuclear microsatellite markers to assess inter- and intra-population gene flow in five natural populations of Luehea divaricata growing in the Pampa biome, in southern Brazil. This species occurs in practically all secondary forests of the Pampa and has recognized ecological significance for these formations. The genetic structuring of the studied populations suggests limited gene dispersal among forest fragments, despite a homogeneous level of migration among populations. Notwithstanding the gene flow among populations, significant SGS is still found in some fragments. Significant spatial genetic structure within population was also found likely as result of limited seed and/or pollen dispersal. The scattered distribution of the populations and their relatively high density seem to limit pollen dispersal. Also seed dispersal by wind is not efficient due to large distances among forest formations. As conservationist actions towards preserving the genetic resources of L. divaricata and the Brazilian Pampa, we suggest the protection of the existing forest formations and the maintenance of the natural expansion of the forests over the grasslands in the biome.





Similar content being viewed by others
References
Backes P, Irgang B (2002) Árvores do Sul. Guia de identificação e interesse Ecológico. As principais espécies nativas sul-brasileiras, Instituto Souza Cruz
Beerli P (2004) Migrate: documentation and program, part of LAMARC Version 2.0. http://evolution.ge.washington.edu/lamarc.html
Beerli P, Felsenstein J (1999) Maximum-likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152:763–773
Carvalho PER (2003) Espécies arbóreas brasileiras ed. Embrapa Informação Tecnológica, Brasília, Brasília
Conson ARO, Ruas EA, Vieira BG, Rodrigues LA, Costa BF, Bianchini E, Prioli AJ, Ruas CF, Ruas PM (2013) Genetic structure of the Atlantic Rainforest tree species Luehea divaricata (Malvaceae). Genetica 141:205–215
Cordeiro JLP, Hasenack H (2009) Cobertura vegetal atual do Rio Grande do Sul. In: Pillar VP, Müller SC, Castilhos ZMS, Jacques AVA (eds) Campos Sulinos: conservação e uso sustentável da biodiversidade. Brasília, MMA, pp 285–299
De Carvalho MCCG, Da Silva DCG, Ruas PM, Medri ME, Ruas EA, Ruas CF (2008) Flooding tolerance and genetic diversity in populations of Luehea divaricata. Biol Plant 52:771–774
De Meester L, Gomez A, Okamura B, Schwenk K (2002) The monopolization hypothesis and the dispersalgene flow paradox in aquatic organisms. Acta Oecol 23:121–135
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131(479–4):91
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Goodnight KF, Queller DC (1999) Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol Ecol 8:1231–1234
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier M-H et al (2006) Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol Ecol 15:559–571
Heuertz M, Vekemans X, Hausman J-F, Palada M, Hardy OJ (2003) Estimating seed versus pollen dispersal from spatial genetic structure in the common ash. Mol Ecol 12:2483–2495
Heywood JS (1991) Spatial analysis of genetic variation in plant populations. Annu Rev Ecol Syst 22:335–355
IBGE (2004) Mapa de Biomas do Brasil. http://www.ibge.gov.br. Accessed 20 Dec 2013
Konovalov DA, Manning C, Henshaw MT (2004) KINGROUP: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol Ecol Notes 4:779–782
Ladizinsky G (1985) Founder effect in crop-plant evolution. Econ Bot 39:191–199
Langella O (2002) Populations (Version 1.2.28). Centre National de la Recherche Scientifique, France
Lemos RPM, D’Oliveira CB, Rodrigues CR, Roesch LFW, Stefenon VM (2014) Modeling distribution of Schinus molle L. in the Brazilian Pampa: insights on vegetation dynamics and conservation of the biome. Ann For Res. doi:10.15287/afr.2014.272
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Moritz C, Faith DP (1998) Comparative phylogeography and the identification of genetically divergent areas for conservation. Mol Ecol 7:419–430
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Nosil P, Egan SP, Funk DJ (2008) Heterogeneous genomic differentiation between walking-stick ecotypes: ‘isolation by adaptation’ and multiple roles for divergent selection. Evolution 62:316–336
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3:93–114
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol 22:5983–5999
Overbeck GE, Muller SC, Fidelis A, Pfadenhauer J, Pillar VD, Blanco CC, Boldrini II, Both R, Forneck ED (2007) Brazil’s neglected biome: the south Brazilian Campos. Perspect Plant Ecol Evol Syst 9:101–116
Paoli AAS (1995) Morfologia e Desenvolvimento de Sementes e Plântulas de Luehea divaricata Mart. et Zucc. (Tiliaceae). Rev Bras Sem 17:120–128
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pillar VD (2003). Dinâmica da expansão florestal em mosaicos de floresta e campos no sul do Brasil [Dynamic of the forest expansion in mosaics of forest and grasslands in South Brazil]. In: Claudino-Sales V (Ed.) Ecossistemas Brasileiros: Manejo e Conservação, Expressão Gráfica e Editora [a Brazilian publisher], Fortaleza [Brazil], pp. 209–216
Pritchard JK, Stephens M, Donnely P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 43:258–275
Roesch LFW, Vieira FCB, Pereira VA, Schünemann AL, Teixeira IF, Senna AJT, Stefenon VM (2009) The Brazilian Pampa: a fragile biome. Diversity 1:182–198
Rohlf FJ (1998) NTSYS-pc: numerical taxonomy and multivariate analysis system (version 2.0). State University of New York, New York
Ruas EA, Conson ARO, Costa BF, Damasceno JO, Rodrigues LA, Reck M, Vieira AOS, Ruas PM, Rua CF (2009) Isolation and characterization of tem microsatellite loci for the tree species Luehea divaricata Mart. (Malvaceae) and intergeneric transferability. Conserv Genet Resour 1:245–248
Schwaegerle KE, Schaal BA (1979) Genetic variability and founder effect in the pitcher plant Sarracenia purpurea L. Evolution 33:1210–1218
Slatkin M (1995) A measure of population division based on microsatellite allele frequencies. Genetics 139:457–462
Sork VL, Nason J, Campbell DR, Fernandez JF (1999) Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol Evol 14:219–224
Stefenon VM, Gailing O, Finkeldey R (2008) The role of gene flow in shaping genetic structures of the subtropical conifer species Araucaria angustifolia. Plant Biol 10:356–364
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935
Wall SBV (2003) Effects of seed size of wind-dispersed pines (Pinus) on secondary seed dispersal and the caching behavior of rodents. Oikos 100:25–34
Wright S (1943) Isolation by distance. Genetics 28:139–156
Wright S (1946) Isolation by distance under diverse systems of mating. Genetics 31:39–59
Acknowledgments
We would like to thank CNPq (Processes 471812/2011-0 and 474758/2012-5) and UNIPAMPA (PROPESQ and PROPG) by the financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Nagel, J.C., Ceconi, D.E., Poletto, I. et al. Historical gene flow within and among populations of Luehea divaricata in the Brazilian Pampa. Genetica 143, 317–329 (2015). https://doi.org/10.1007/s10709-015-9830-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-015-9830-9