Abstract
Transposable elements are important factors driving plant genome evolution. Upon their mobilization, novel insertion polymorphisms are being created. We investigated differences in copy number and insertion polymorphism of a group of Mariner-like transposable elements Vulmar and related VulMITE miniature inverted-repeat transposable elements (MITEs) in species representing subfamily Betoideae. Insertion sites of these elements were identified using a modified transposon display protocol, allowing amplification of longer fragments representing regions flanking insertion sites. Subsequently, a subset of TD fragments was converted into insertion site-based polymorphism (ISBP) markers. The investigated group of transposable elements was the most abundant in accessions representing the section Beta, showing intraspecific insertion polymorphisms likely resulting from their recent activity. In contrast, no unique insertions were observed for species of the genus Beta section Corollinae, while a set of section-specific insertions was observed in the genus Patellifolia, however, only two of them were polymorphic between P. procumbens and P. webbiana. We hypothesize that Vulmar and VulMITE elements were inactivated in the section Corollinae, while they remained active in the section Beta and the genus Patellifolia. The ISBP markers generally confirmed the insertion patterns observed with TD markers, including presence of distinct subsets of TE insertions specific to Beta and Patellifolia.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transposable elements (TEs) constitute a significant portion of plant genomes. Their content in Angiospermae genomes varies from ca. 10% in Arabidopsis (Arabidopsis Genome Initiative 2000) to ca. 85% in maize genome (Schnable et al. 2009). TEs are capable of changing their localization in the genome and their activity may alter gene structure and regulatory functions or cause genome rearrangements (Bennetzen 2000). On the basis of their mechanism of transposition, TEs are divided into two classes. Class I elements include retrotransposons which transpose via an RNA intermediate. Subsequently, the RNA is reverse-transcribed and the DNA copy is integrated into a new genomic location. The ‘copy and paste’ transposition is replicative and each transposition event leads to increase of the TE copy number. In contrast, Class II elements—DNA transposons, are mobilized on the basis of a ‘cut and paste’ mechanism, i.e. their mobilization requires physical excision from a donor site and reintegration into a new acceptor site. Autonomous elements contain functional open reading frame(s) that encode the products required for transposition. Deletions in the coding regions are typical for non-autonomous elements, which nevertheless may be activated in trans, provided that the required transposition machinery encoded by a related autonomous element is present (Wessler 2006).
Autonomous elements and their non-autonomous derivatives are grouped into families defined by DNA sequence conservation (Wicker et al. 2007). TE families differ in the TE copy number, which can be exceptionally high for some retrotransposon families. Also, a particular group of TEs, called miniature inverted-repeat transposable elements (MITEs) can reach hundreds to thousands copies per haploid genome (Feschotte et al. 2002a). MITEs are small, up to 600 bp, DNA transposons lacking any coding capacity. They were identified in several plant species, including maize (Bureau and Wessler 1992), rice (Bureau and Wessler 1994a, b), Arabidopsis (Casacuberta et al. 1998, Feschotte et al. 2003), carrot (Grzebelus and Simon 2009), and Medicago (Grzebelus et al. 2009). Because of the shared structural features, it was proposed that MITEs were reminiscent of non-autonomous elements. It is likely that the high copy number of MITEs is an effect of their in trans activation by related autonomous elements (Feschotte et al. 2002b).
A Vulmar/VulMITE group of TEs identified in Beta could be an example of such interrelationship. Vulmar1 is a TE belonging to the Tc1/mariner superfamily and a representative of the Vulmar family. Vulmar1 is 3,909 bp-long, non-autonomous due to frameshifts in the transposase gene. However, many open reading frames coding for mariner-like transposases were identified in beet, some of them likely representing autonomous elements, including those from the Vulmar family (Jacobs et al. 2004). Menzel et al. (2006) reported on three MITE families, named VulMITE I, II, and III, present in B. vulgaris and related to the Vulmar elements. While VulMITE I were typical MITEs with length of ca. 300 bp, the other two families were ca. 1 kb-long. Sequence similarity of the MITEs and Vulmar1 was restricted to less than 200 bp 5′ and 3′ terminal regions. Menzel et al. (2006) also indicated that insertions of VulMITE I elements were polymorphic in Beta, resulting from transposition events, likely in the course of beet domestication.
A successful transposition event results in a novel insertion polymorphism. It can be detected by a range of techniques, including transposon display (TD). Transposon display is a modification of the AFLP system, in which subterminal fragments of transposons are used as target sequences. MITE-based transposon display was used to detect insertion polymorphisms in maize (Casa et al. 2000), rice (Park et al. 2003, Jiang et al. 2003, Kwon et al. 2006), and carrot (Grzebelus and Simon 2009). TE insertion polymorphisms can be applied to elucidate interspecific evolutionary relationships (Ray 2007). In order to investigate such relationships in Betoideae, we used TD approach to study Vulmar/VulMITE insertion polymorphisms.
Historically, all investigated species, representing family Chenopodiaceae/Amaranthaceae, were grouped in the genus Beta, which was divided into four sections Ulbrich (1934). The section Procumbentes, grouping B. procumbens, B. patellaris, and B. webbiana was eventually removed from the genus Beta and the species were included in the genus Patellifolia (Scott et al. 1977). According to the current USDA/ARS NPGS GRIN taxonomy (www.ars-grin.gov/cgi-bin/npgs/html/gnlist.pl?2303, August 17, 2011), both Beta and Patellifolia are classified in subfamily Chenopodioideae, tribe Beteae. However, Hohmann et al. (2006) placed them in subfamily Betoideae, and provided molecular evidence for further division of Betoideae into Beteae comprising all Beta species, and a sister group, Hablitzieae, including Patellifolia. Within Beteae, two sections are recognized—section Beta, comprising B. vulgaris and B. macrocarpa, and section Corollinae, comprising B. corolliflora, B. lomatogona, B. macrorhiza, B. trygina, and B. nana (Kadereit et al. 2006).
Here, we describe differences in the copy number and insertion polymorphism of Vulmar/VulMITE transposons among Beta and Patellifolia species, likely reflecting differences in evolutionary dynamics of these elements.
Materials and methods
Plant material and DNA extraction
Two male sterile and two corresponding maintainer (Owen type) sugar beet breeding stocks (KHBC Straszków, Poland), together with four wild Beta species, i.e. B. vulgaris subsp. maritima (section Beta), B. corolliflora, B. lomatogona, B. macrorhiza (section Corollinae), and two Patellifolia species, i.e. P. procumbens, and P. webbiana, were used for Vulmar/VulMITE transposon display (VMTD). Each accession was represented by two plants. Insertion site-based polymorphism (ISBP) markers were applied to the above materials and few additional sugar beet and beetroot stocks (Table 1). DNA was extracted with DNeasy Mini Kit (Qiagen) as described in the manufacturer protocol.
Vulmar/VulMITE transposon display (VMTD)
Approximately 300 ng of DNA samples were digested in 37°C for 3 h in 10 μl reaction mixture with 5 u NdeI (Fermentas). NdeI-compatible splinkerette adaptors were ligated to the restriction fragments according to the protocol of Grzebelus et al. (2007). Preamplification was performed with a primer VMtd1 (5′ GCCTAGGAGTCCGTTTTAATCAC 3′), specific to 5′ subterminal region of Vulmar1 and VulMITE elements (Menzel et al. 2006), and with an adaptor-specific primer tdP1 (5′ CGAATCGTAACCGTTCGTACGAGAA 3′) in 10 μl reaction mixture containing 0.5 μM of each primer, 0.5 U of Taq polymerase (Fermentas), 250 μM dNTP, and 1.5 mM MgCl2 in the PCR buffer provided by the manufacturer. The following cycling conditions were applied: initial denaturation at 94°C/3 min., then 40 cycles of 94°C/30 s, 55°C/30 s, and 68°C/60 s, final elongation at 68°C/5 min. The reaction mixture was diluted 1:20 and used as a template in selective amplification. Selective amplifications were performed with a nested Vulmar1- and VulMITE-specific primer VMtd2 (5′ CTAGGAGTCCGTTTTAATCACAATG 3′) and a nested adaptor-specific primer tdNdeI(NN) (5′ TCCAACGAGCCAAGGTATGNN 3′), where ‘NN’ stands for the two selective nucleotides attached to the 3′ end. Eight combinations of the two selective nucleotides were used, i.e. AC, AG, CA, CT, GA, GT, TC, and TG. The reaction was set up in 10 μl volume containing 0.5 μM of each primer, 0.5 U of Taq polymerase (Fermentas), 1.5 mM MgCl2, 250 μM dNTP, and 1xPCR buffer. Cycling conditions for selective amplification were set according to Grzebelus et al. (2007). TD amplicons were separated in 1% agarose gels and visualized with ethidium bromide.
Cloning and sequencing of PCR products
Target bands were cut from gels and purified with WIZARD SV Gel and PCR Clean-Up System (Promega) following the instructions provided by the manufacturer. Purified DNA fragments were ligated into pGem-T (Promega), cloned, and sequenced using GenomeLab DTCS Quick Start Kit (Beckman Coulter) and separated in CEQ 8000 (Beckman Coulter) according to the manufacturer protocol. Sequences were analyzed with BioEdit (Hall 1999). Fourteen sequences representing flanking regions of TE insertions longer than 200 bp were deposited in GenBank (accession numbers: JF521550-JF521563).
Site-specific PCR
In order to convert VMTD polymorphisms into ISBP markers, primers anchored in the region flanking TE insertion sites were designed using Primer3 for the sequenced VMTD fragments (Supplementary Table 1). These primers were used for PCR in combination with the VMtd2 primer. DNA amplification was carried out in 10 μl reaction mixture containing 0.5 μM of each primer, 0.5 U of Taq polymerase (Fermentas), 250 μM dNTP, 2 mM MgCl2. The following cycling conditions were applied: initial denaturation at 94°C/2 min., then 35 cycles of 94°C/30 s, 53 or 58°C/30 s, and 68°C/2 min., final elongation at 68°C/5 min. Products were separated in 1% agarose gels and visualized with ethidium bromide.
Analysis of genetic diversity
Presence or absence of VMTD bands was scored as 1 or 0, respectively and a binary matrix was created. Pairwise genetic distances were calculated as proposed for binary data by Huff et al. (1993), i.e. any comparison with the same state yielded a value of 0, while different states yielded a value of 1. Analysis of molecular variance (AMOVA) was performed with two sources of variation—stocks and plants. Test for statistical significance of components was based on 1,000 permutations. Principal Coordinates Analysis (PCoA) was performed using a Distance-Standardized method. All calculations were carried out in GeneAlex 6.3 (Peakall and Smouse 2006).
Results
Vulmar/VulMITE transposon display
Eight combinations of selective nucleotides produced the total of 96 VMTD bands. The number of fragments ranged from 7 to 16, for selective nucleotide combinations AC and GA, respectively (Table 2). The average number of bands per one reaction was 11.9, only slightly lower than that obtained using a similar system based on Krak MITEs in carrot (Grzebelus and Simon 2009). Our protocol was a modification of the latter TD system. We replaced MseI—a four-cutter, with NdeI which is a six-cutter, which allowed production of longer TD fragments which were separated on agarose gels. The original TD protocol yielded products that were on average ca. 100 bp-long (Grzebelus and Simon 2009), while the average length of VMTD products was ca. 350 bp. Nearly all VMTD fragments (97.8%) were polymorphic among the investigated species.
Distribution of Vulmar/VulMITE insertions in Betoideae
AMOVA partitioned 99% of molecular variance to differences among stocks, while only the remaining 1% could be attributed to the intra-stock variability. On the basis of the observed insertion polymorphism, the investigated Betoideae species could be divided into four distinct groups representing (1) sugar beet, (2) B. vulgaris subsp. maritima, (3) section Corollinae: B. corolliflora, B. lomatogona, B. macrorhiza, and (4) the genus Patellifolia (Fig. 1). For the wild species, the results were consistent with the current taxonomy of Betoideae, while separate positions of the cultivated and the wild B. vulgaris could be attributed to mobilization of the TEs, as discussed below.
Principal coordinates analysis (PCoA) of Betoideae based on VMTD polymorphisms. Two individuals represent each stock. Individuals with identical VMTD scores occupy the same position on the graph. Plant stock codes are given according to Table 1
Species belonging to section Corollinae were characterized by a low number of identified insertions, all but one of them being shared with species representing other sections. A much higher number of insertions, i.e. 50 and 19, were specific to section Beta and genus Patellifolia, respectively (Fig. 2). Of the 50 insertion sites present exclusively in the section Beta, 27 and 14 bands were unique to sugar beet and B. vulgaris subsp. maritima, respectively. In contrast, the two investigated Patellifolia species shared a set of 35 insertion sites, while only two TD bands were present in P. procumbens and absent in P. webbiana.
Graph illustrating relationships among the investigated Beta species with regard to the number of shared and unique VulMITE I insertions. Section Beta [Beta]—sugar beet, B. vulgaris subsp. maritima [Bvm], section Corollinae [Cor]—B. corolliflora [Bc], B. lomatogona [Bl], B. macrorhiza [Bm], and genus Patellifolia [Pat]—(P. procumbens [Pp], P. webbiana [Pw])
Conversion of VMTD polymorphisms into ISBP markers
To further elaborate the nature of polymorphisms identified by VMTD we sequenced 34 VMTD fragments, including nine groups (comprising two to five sequences) of co-migrating fragments amplified in plants from different accessions. Sixteen and seven fragments selected for sequencing originated from sugar beet and B. vulgaris subsp. maritima, respectively, while five and six were chosen for Corollinae and Patellifolia, respectively. Sequences of the co-migrating products always matched each other, so we obtained 20 sequences representing regions flanking Vulmar/VulMITE insertion sites. All sequenced TD fragments showed similarity to the terminal part of Vulmar and VulMITE immediately following the anchor site of the VMtd2 primer, indicating that the fragments were specifically derived from regions bearing TE insertions.
Expected amplification products were obtained for 17 of the 20 sequenced insertion sites. Two PCRs produced complex banding patterns, likely indicating that the primer specific to the flanking region was anchored in the repetitive DNA, while the remaining one did not yield any products. Distribution of site-specific bands generally matched that of the original VMTD products (Fig. 3). In two cases (Bv6 and Bv14), a discrepancy between TD and ISBP scores was observed, likely resulting from scoring different TD bands of similar size as one product (Supplementary Table 2). Thirteen ISBP markers were used to screen a larger genepool representing cultivated beets. As expected, four of them, two originating from Patellifolia and two present exclusively in B. vulgaris subsp. maritima, were absent both in sugar beet and beetroot. The remaining nine ISBP markers segregated in the larger cultivated beet genepool (Table 3). Grouping of the investigated species based on the 13 ISBP polymorphisms resembled that obtained for the VMTD markers (Fig. 4). While both Patellifolia species and three representatives of Procumbentes showed distinct and uniform polymorphism patterns, a substantial diversity was observed in B. vulgaris. Wild B. vulgaris subsp. maritima differed from the cultivated materials, while sugar beet and beetroot formed two only marginally overlapping groups.
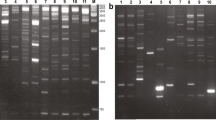
An example of co-segregation of the VMTD markers obtained with CT selective nucleotides and the derived ISBP markers Bv7 and Bp1. VMTD profile (a), arrows indicate localization of sequenced fragments. Amplification of ISBP markers: sugar beet-specific Bv7 (b) and Patellifolia-specific Pp1 (c), M molecular size marker
Principal coordinates analysis (PCoA) of Betoideae based on ISBP polymorphisms. Two to four individuals represent each stock. Individuals with identical ISBP scores occupy the same position on the graph. Plant stock codes are given according to Table 1
Discussion
VMTD protocol allowed generation of longer fragments, which in turn facilitated the design of primers anchored in regions flanking Vulmar/VulMITE insertions. We used agarose gels for separation of the TD amplicons, which simplified the detection of polymorphisms. However, it came at a cost of a lower number of bands that could have been identified in one reaction. Other MITE-based TD systems were more efficient and produced from 42 to 77 bands for Heartbreaker in maize (Casa et al. 2000) and 51 for mPing in rice (Naito et al. 2006). Sequences of selected VMTD fragments showed that all of them were derived from insertion sites. Thus, the technique allowed efficient identification of regions flanking TE insertions. The DcMaster TD system in carrot, similar to here reported VMTD, produced ca. 10% of non-specific fragments (Grzebelus et al. 2007).
It should be stressed that the observed VMTD polymorphism, especially on the interspecific level, may not result solely from TE insertion polymorphism, but also from single nucleotide polymorphisms in restriction sites or rearrangements of primer annealing sites. It is not possible to directly estimate the relative contribution of these factors to the overall level of the revealed interspecific differences. Two interrelated factors may have an effect on it, i.e. the distance between species, and the age of an insertion. In relatively distant species, common insertions that can be identified are old, thus there is more chance that the TE itself or the region flanking the insertion has been rearranged. As a result, the observed TD polymorphism may not reflect the actual TE presence/absence, but rather the inability to produce same-sized TD bands for both species. There were attempts to use TE-based platforms as tools for phylogenetic analysis (Nagy et al. 2006; Moisy et al. 2008), however, that problem was never addressed. On the other hand, all differences among the sugar beet stocks should in principle be attributed to the activity of the transposable elements, as transposition seems to be the most likely cause of TD polymorphisms in otherwise very similar genomes.
The data provided the opportunity to hypothesize about the activity of Vulmar and VulMITE in the course of evolution of Betoideae. We conclude that these elements were not active in section Corollinae and the shared insertions present in B. corolliflora, B. lomatogona, and B. macrorhiza were already present in the common ancestor of all Beta species. In contrast, a substantial insertion polymorphism was observed in different sugar beet stocks, corroborating observations of Menzel et al. (2006). The highest copy number of the investigated elements was observed in sugar beet and B. vulgaris subsp. maritima. The identified intraspecific insertion polymorphism suggests that Vulmar and/or at least one of VulMITE families was active in these subspecies, most likely also in the period of beet domestication. Results reported by Menzel et al. (2006) suggest that VulMITE I family could be responsible for that recent burst of avtivity. Similar bursts of MITE activity was reported as an effect of rice domestication (Naito et al. 2006), while Grzebelus et al. (2009) reported on a possibility of periodical MITE mobilization in domesticated M. truncatula.
Some Vulmar/VulMITE elements were active also in Patellifolia, resulting in a subset of novel insertions not present in any Beta species. It has been debated if P. procumbens and P. webbiana should be regarded as separate species (Curtis 1968). In the present study both species shared almost the same set of insertion sites, which supported previous reports on a very limited level of interspecific variability between P. procumbens and P. webbiana (Wagner et al. 1989; Mita et al. 1991). In terms of TE dynamics in Patellifolia, our results indicated that unlike B. vulgaris, their mobilization was much more limited, as there were very little differences between the two investigated species, P. procumbens and P. webbiana as compared to any pair of accessions of B. vulgaris. The divergence time of Betoideae and Hablitzieae was estimated to be 38.4–27.5 Mya (Hohmann et al. 2006), which suggests that the group of Vulmar/VulMITE elements consists of families that have been active in different evolutionary periods. Possibly, different families were active in Beta and Patellifolia, as we observed distinct subsets of insertions differentiating the two genera, which was confirmed by ISBP markers. This could be experimentally verified by cloning and sequencing full-length TEs from the diagnostic sites. Detailed investigation on the activity periods of Vulmar and VulMITE elements could help develop tools for TE insertion-based phylogenetic analysis of Betoideae. That strategy was successfully applied, e.g. the use of SINEs for studying phylogeny of primates (reviewed in Ray 2007). MITEs are a good source of phylogenetic information in plants, owing to their abundance and small size. However, as they derive from Class II, they are capable of excision and sequence verification of insertion sites, identifying footprints created upon excision might be required.
References
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Bennetzen JL (2000) Transposable element contribution to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bureau TE, Wessler SR (1992) Tourist: a large family of small inverted repeat elements frequently associated with maize genes. Plant Cell 4:1283–1294
Bureau TE, Wessler SR (1994a) Mobile inverted-repeat elements of the Tourist family are associated with the genes of many cereal grasses. Proc Natl Acad Sci USA 91:1411–1415
Bureau TE, Wessler SR (1994b) Stowaway: a new family of inverted repeat elements associated with genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6:907–916
Casa AM, Brouwer C, Nagel A, Wang L, Zhang Q, Kresovich S, Wessler SR (2000) The MITE family Heartbreaker (Hbr): molecular markers in maize. Proc Natl Acad Sci USA 97:10083–10089
Casacuberta E, Casacuberta JM, Puigdomenech P, Monfort A (1998) Presence of miniature inverted-repeat transposable elements (MITEs) in the genome of Arabidopsis thaliana: characterization of the Emigrant family of elements. Plant J 16:79–85
Curtis GJ (1968) Observations on fruit shape and other characters in the species of the section Patellares:genus Beta. Euphytica 17:485–491
Feschotte C, Jiang N, Wessler SR (2002a) Plant transposable elements: where genetics meets genomics. Nature Rev Genet 3:329–341
Feschotte C, Zhang X, Wessler SR (2002b) Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: L Craig, R Craigie, M Gellert, A Lambowitz (eds) Mobile DNA II. ASM Press, Washington, DC, pp 1147–1158
Feschotte C, Swamy L, Wessler SR (2003) Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway miniature inverted repeat transposable elements (MITEs). Genetics 163:747–758
Grzebelus D, Simon PW (2009) Diveristy of DcMaster-like elements of the PIF/Harbinger superfamily in the carrot genome. Genetica 135:347–353
Grzebelus D, Jagosz B, Simon PW (2007) The DcMaster transposon display maps polymorphic insertion sites in the carrot (Daucus carota L.) genome. Gene 390:67–74
Grzebelus D, Gladysz M, Macko-Podgorni A, Gambin T, Golis B, Rakoczy R, Gambin A (2009) Population dynamics of miniature inverted-repeat transposable elements (MITEs) in Medicago truncatula. Gene 448:214–220
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hohmann S, Kadereit JW, Kadereit G (2006) Understanding Mediterranean-Californian disjunctions: molecular evidence from Chenopodiaceae-Betoideae. Taxon 55:67–78
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among natural populations of outcrossing buffalograss Buchloe dactyloides (Nutt) Engelm. Theor Appl Genet 86:927–934
Jacobs G, Dechyeva D, Menzel G, Dombrowski C, Schmidt T (2004) Molecular characterization of Vulmar1, a complete mariner transposon of sugar beet and diversity of mariner- and En/Spm-like sequences in the genus Beta. Genome 47:1192–1201
Jiang N, Bao Z, Zhang Z, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Kadereit G, Hohmann S, Kadereit JW (2006) A synopsis of Chenopodiaceae subfam. Betoideae and notes on the taxonomy of Beta. Willdenowia 36:9–19
Kwon SJ, Hong SW, Son JH, Lee JK, Cha YS, Eun MY, Kim NS (2006) CACTA and MITE transposon distributions on a genetic map of rice using F15 RILs derived from Milyang 23 and Gihobyeo hybrids. Mol Cell 21:360–366
Menzel G, Dechyeva D, Keller H, Lange C, Himmelbauer H, Schmidt T (2006) Mobilization and evolutionary history of miniature inverted-repeat transposable elements (MITEs) in Beta vulgaris L. Chromosome Res 14:831–844
Mita G, Dani M, Casciari P, Pasquali A, Selva E, Minganti C, Piccardi P (1991) Assessment of the degree of genetic variation in beet based on RFLP analysis and the taxonomy of Beta. Euphytica 55:1–6
Moisy C, Garrison KE, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1/copia-like retrotransposon families of the grapevine genome. BMC Genom 9:469
Nagy ED, Molnár I, Schneider A, Kovács G, Molnár-Láng M (2006) Characterization of chromosome-specific S-SAP markers and their use in studying genetic diversity in Aegilops species. Genome 49:289–296
Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR (2006) Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA 103:17620–17625
Park KC, Kim NH, Cho YS, Kang KH, Lee JK, Kim NS (2003) Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor Appl Genet 107:203–209
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Ray DA (2007) SINEs of progress: mobile element applications to molecular ecology. Mol Ecol 16:19–33
Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326:1112–1115
Scott AJ, Ford-Lloyd BV, Williams JT (1977) Patellifolia, nomen ovum (Chenopodiaceae). Taxon 26:284
Ulbrich E (1934) Chenopodiaceae. In: Engler A, Prantl K (eds) Die natürlichen Pflanzenfamilien, 2nd edn. Leipzig, pp 379–584
Wagner H, Gimbel EM, Wricke G (1989) Are Beta procumbens Chr. Sm. and Beta webbiana Moq. different species? Plant Breed 102:17–21
Wessler SR (2006) Eukaryotic transposable elements: teaching old genomes new tricks. In: Caporale L (ed) The implicit genome. Oxford University Press, Oxford, pp 138–165
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Acknowledgments
The research was funded by the Polish Ministry of Science and Higher Education, grant no. PBZ-MNiSW-2/3/2006/35 awarded to MS.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Grzebelus, D., Stawujak, K., Mitoraj, J. et al. Dynamics of Vulmar/VulMITE group of transposable elements in Chenopodiaceae subfamily Betoideae . Genetica 139, 1209–1216 (2011). https://doi.org/10.1007/s10709-011-9622-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-011-9622-9