Abstract
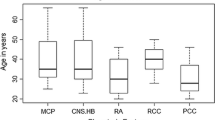
The data in genotype-phenotype correlation in Indian von Hippel–Lindau (VHL) patients is limited. We have retrospectively studied 31 genetically proven VHL patients with pheochromocytoma/paraganglioma (PCC/PGL) from families and have reviewed the World literature on PCC/PGL in patients with large VHL deletions. Three patients had large deletions and 28 patients had other mutations [missense mutations in 25, 3 bp deletion in 2 and single bp duplication in one]. Unilateral PCC were significantly more common in patients with large VHL deletions whereas multiple PCC (bilateral PCC or PCC + sympathetic PGL) were significantly more common in those with other mutations. World literature review confirmed the rarity of PCC/PGL in patients with large deletions and we report the first definitive case of PCC associated with complete VHL deletion. Pancreatic neuroendocrine tumours were more common, often metastatic and the most common cause of death in our cohort. Our study had eight parent off-spring pairs from five families. The off-springs were significantly younger at presentation and had significantly higher number of PCC/PGL. In conclusion, PCC/PGL are rare in patients with large VHL deletions and if occur are most likely to be solitary. Patients with bilateral PCC or multifocal PCC/PGL are least likely to have large VHL deletions. Our study also provides additional evidence for existence of the phenomenon of anticipation in VHL syndrome.
Similar content being viewed by others
References
Maher ER, Iselius L, Yates JR et al (1991) Von Hippel–Lindau disease: a genetic study. J Med Genet 28(7):443–447
Ong KR, Woodward ER, Killick P, Lim C, Macdonald F, Maher ER (2007) Genotype–phenotype correlations in von Hippel–Lindau disease. Hum Mutat 28(2):143–149
Maher ER, Yates JR, Harries R et al (1990) Clinical features and natural history of von Hippel–Lindau disease. Q J Med 77(283):1151–1163
Gläsker S, Neumann HPH, Koch CA, Vortmeyer AO (2000) Von Hippel–Lindau Disease. In: De Groot LJ, Chrousos G, Dungan K, Feingold KR, Grossman A, Hershman JM, Koch C, Korbonits M, McLachlan R, New M, Purnell J, Rebar R, Singer F, Vinik A (eds) Endotext [Internet]. MDText.com, Inc., South Dartmouth, MA
Chou A, Toon C, Pickett J, Gill AJ (2013) Von Hippel–Lindau syndrome. Front Horm Res 41:30–49
Gossage L, Eisen T, Maher ER (2015) VHL, the story of a tumour suppressor gene. Nat Rev Cancer 15(1):55–64
McNeill A, Rattenberry E, Barber R, Killick P, MacDonald F, Maher ER (2009) Genotype–phenotype correlations in VHL exon deletions. Am J Med Genet A 149A(10):2147–2151
Pandit R, Khadilkar K, Sarathi V, Kasaliwal R et al (2016) Germline mutations and genotype–phenotype correlation in Asian Indian patients with pheochromocytoma and paraganglioma. Eur J Endocrinol 175(4):311–323
Vikkath N, Valiyaveedan S, Nampoothiri S et al (2015) Genotype–phenotype analysis of von Hippel–Lindau syndrome in fifteen Indian families. Fam Cancer 14(4):585–594
Ebenazer A, Rajaratnam S, Pai R et al (2013) Detection of large deletions in the VHL gene using a real–time PCR with SYBR green. Fam Cancer 12(3):519–524
Janavicius R, Adomaitis R, Jankevicius F, Griskevicius L (2009) Extremely low risk of pheochromocytomas in complete VHL gene deletion cases. Hum Mutat 30(9):1365–1366. https://doi.org/10.1002/humu.21050
Untergasser A, Nijveen H, Rao X et al (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35:W71–W74
Schwarz JM, Cooper DN, Schuelke M et al (2014) MutationTaster2: mutation prediction for the deep–sequencing age. Nat Methods 11:361–362
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding nonsynonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081
Stenson PD, Mort M, Ball EV et al (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 133:1–9
Beroud C, Joly D, Gallou C et al (1998) Software and database for the analysis of mutations in the VHL gene. Nucleic Acids Res 26:256–258
Gossage L, Pires DE, Olivera–Nappa Á (2014) An integrated computational approach can classify VHL missense mutations according to risk of clear cell renal carcinoma. Hum Mol Genet 15(22):5976–5988
Nordstrom–O’Brien M, van der Luijt RB, van Rooijen E et al (2010) Genetic analysis of von Hippel–Lindau disease. Hum Mutat 31(5):521–537
Wong M, Chu YH, Tan HL et al (2016) Clinical and molecular characteristics of East Asian patients with von Hippel–Lindau syndrome. Chin J Cancer 35:79
Crossey PA, Richards FM, Foster K, Green JS et al (1994) Identification of intragenic mutations in the von Hippel–Lindau disease tumour suppressor gene and correlation with disease phenotype. Hum Mol Genet 3(8):1303–1308
Chen F, Kishida T, Yao M, Hustad T, Glavac D et al (1995) Germline mutations in the von Hippel–Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat 5(1):66–75
Franke G, Bausch B, Hoffmann MM et al (2009) Alu Alu recombination underlies the vast majority of large VHL germline deletions: Molecularcharacterization and genotype–phenotype correlations in VHL patients. Hum Mutat 30(5):776–786
Hes F, Zewald R, Peeters T, Sijmons R et al (2000) Genotype–phenotype correlations in families with deletions in the von Hippel–Lindau (VHL) gene. Hum Genet 106(4):425–431
Cybulski C, Krzystolik K, Murgia A, Górski B, Debniak T et al (2002) Germline mutations in the von Hippel–Lindau (VHL) gene in patients from Poland: disease presentation in patients with deletions of the entire VHL gene. J Med Genet 39(7):E38
Rocha JC, Silva RL, Mendonça BB, Marui S et al (2003) High frequency of novel germline mutations in the VHL gene in the heterogeneous population of Brazil. J Med Genet 40(3):e31
Maranchie JK, Afonso A, Albert PS, Kalyandrug S et al (2004) Solid renal tumor severity in von Hippel–Lindau disease is related to germline deletion length and location. Hum Mutat 23(1):40–46
Hes FJ, van der Luijt RB, Janssen AL et al (2007) Frequency of Von Hippel–Lindau germline mutations in classic and non–classic Von Hippel–Lindau disease identified by DNA sequencing, Southern blot analysis and multiplex ligation–dependent probe amplification. Clin Genet 72(2):122–129
Cascón A, Escobar B, Montero–Conde C et al (2007) Loss of the actin regulator HSPC300 results in clear cell renal cell carcinoma protection in Von Hippel–Lindau patients. Hum Mutat 28(6):613–621
Huang JS, Huang CJ, Chen SK, Chien CC (2007) Associations between VHL genotype and clinical phenotype in familial von Hippel–Lindau disease. Eur J Clin Invest 37(6):492–500
Cho HJ, Ki CS, Kim JW (2009) Improved detection of germline mutations in Korean VHL patients by multiple ligation–dependent probe amplification analysis. J Korean Med Sci 24(1):77–83
Gergics P, Patocs A, Toth M, Igaz P, Szucs N, Liko I et al (2009) Germline VHL gene mutations in Hungarian families with von Hippel–Lindau disease and patients with apparently sporadic unilateral pheochromocytomas. Eur J Endocrinol 161(3):495–502
Gomy I, Molfetta GA, de Andrade Barreto E et al (2010) Clinical and molecular characterization of Brazilian families with von Hippel–Lindau disease: a need for delineating genotype–phenotype correlation. Fam Cancer 9(4):635–642
Wu P, Zhang N, Wang X, Ning X, Li T, Bu D et al (2012) Family history of von Hippel–Lindau disease was uncommon in Chinese patients: suggesting the higher frequency of de novo mutations in VHL gene in these patients. J Hum Genet 57(4):238–243
Wang X, Zhang N, Ning X, Li T, Wu P, Peng S et al (2014) Higher prevalence of novel mutations in VHL gene in Chinese Von Hippel–Lindau disease patients. Urology 83(3):675-e1
Hwang S, Ku CR, Lee JI, Hur KY, Lee MS, Lee CH, Koo KY et al (2014) Germline mutation of Glu70Lys is highly frequent in Korean patients with von Hippel–Lindau (VHL) disease. J Hum Genet 59(9):488–493. https://doi.org/10.1038/jhg.2014.61
Kruizinga RC, Sluiter WJ, de Vries EG, Zonnenberg BA, Lips CJ et al (2013) Calculating optimal surveillance for detection of von Hippel–Lindau–related manifestations. Endocr Relat Cancer 21(1):63–71
Wittström E, Nordling M, Andréasson S (2014) Genotype–phenotype correlations, and retinal function and structure in von Hippel–Lindau disease. Ophthalmic Genet 35(2):91–106
Krzystolik K, Jakubowska A, Gronwald J et al (2014) Large deletion causing von Hippel–Lindau disease and hereditary breast cancer syndrome. Hered Cancer Clin Pract 12(1):16
Ning XH, Zhang N, Li T, Wu PJ, Wang X, Li XY et al (2014) Telomere shortening is associated with genetic anticipation in Chinese Von Hippel–Lindaudisease families. Cancer Res 74(14):3802–3809
Lee JS, Lee JH, Lee KE, Kim JH, Hong JM, Ra EK et al (2016) Genotype–phenotype analysis of von Hippel–Lindau syndrome in Korean families: HIFα bindingsite missense mutations elevate age–specific risk for CNS hemangioblastoma. BMC Med Genet 17(1):48
Peng S, Shepard MJ, Wang J, Li T, Ning X et al (2017) Genotype–phenotype correlations in Chinese von Hippel–Lindau disease patients. Oncotarget. https://doi.org/10.18632/oncotarget.16594
Igarashi H, Ito T, Nishimori I et al (2014) Pancreatic involvement in Japanese patients with von Hippel–Lindau disease: results of a nationwide survey. J Gastroenterol 49(3):511–516
Binkovitz LA, Johnson CD, Stephens DH (1990) Islet cell tumors in von Hippel–Lindau disease: increased prevalence and relationship to the multiple endocrine neoplasias. AJR Am J Roentgenol 155(3):501–505
Charlesworth M, Verbeke CS, Falk GA et al (2012) Pancreatic lesions in von Hippel–Lindau disease? A systematic review and meta–synthesis of the literature. J Gastrointest Surg 16(7):1422–1428
Blansfield JA, Choyke L, Morita SY, Choyke PL, Pingpank JF et al (2007) Clinical, genetic and radiographic analysis of 108 patients with von Hippel–Lindau disease (VHL) manifested by pancreatic neuroendocrine neoplasms (PNETs). Surgery 142(6):814–818
Richards FM, Payne SJ, Zbar B, Affara NA et al (1995) Molecular analysis of de novo germline mutations in the von Hippel–Lindau disease gene. Hum Mol Genet 4(11):2139–2143
Oberlé I, Rousseau F, Heitz D, Kretz C, Devys D et al (1991) Instability of a 550 bp DNA segment and abnormal methylation in fragile X syndrome. Science 252(5009):1097–1102
La Spada AR, Wilson EM, Lubahn DB et al (1991) Androgen receptor gene mutations in X–linked spinal and bulbar muscular atrophy. Nature 352(6330):77–79
Mahadevan M, Tsilfidis C, Sabourin L, Shutler G et al (1992) Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 255(5049):1253–1255
Ranen NG, Stine OC, Abbott MH et al (1995) Anticipation and instability of IT–15 (CAG)n repeats in parent–offspring pairs with Huntington disease. Am J Hum Genet 57(3):593–602
Tabori U, Nanda S, Druker H, Lees J, Malkin D (2007) Younger age of cancer initiation is associated with shorter telomere length in Li–Fraumeni syndrome. Cancer Res 67(4):1415–1418
Martinez–Delgado B, Yanowsky K, Inglada–Perez L et al (2011) Genetic anticipation is associated with telomere shortening in hereditary breast cancer. PLoS Genet 7(7):e1002182
Seguí N, Pineda M, Guinó E, Borràs E, Navarro M et al (2013) Telomere length and genetic anticipation in Lynch syndrome. PLoS ONE 8(4):e61286
Nilbert M, Timshel S, Bernstein I, Larsen K (2009) Role for genetic anticipation in Lynch syndrome. J Clin Oncol 27(3):360–364
Funding
The funding was provided by Scientific and Engineering Research Board (SERB), Department of Science and Technology, Government of India (Grant No. # SB/SO/HS/041/2013).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Lomte, N., Kumar, S., Sarathi, V. et al. Genotype phenotype correlation in Asian Indian von Hippel–Lindau (VHL) syndrome patients with pheochromocytoma/paraganglioma. Familial Cancer 17, 441–449 (2018). https://doi.org/10.1007/s10689-017-0058-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-017-0058-y