Abstract
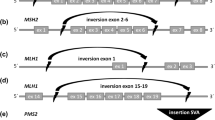
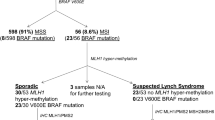
Lynch syndrome (LS) is an autosomal dominant disorder, with high penetrance that affects approximately 3% of the cases of colorectal cancer. Affected individuals inherit germline mutations in genes responsible for DNA mismatch repair, mainly at MSH2, MLH1, MSH6 and PMS2. The molecular screening of these individuals is frequently costly and time consuming due to the large size of these genes. In addition, PMS2 mutation detection is often a challenge because there are 16 different pseudogenes identified until now. In the present work we evaluate a molecular screening strategy based in next generation sequencing (NGS) in order to optimize the mutation detection in LS patients. We established 16 multiplex PCRs for MSH2, MSH6 and MLH1 and 5 Long-Range PCRs for PMS2, coupled with NGS. The strategy was validated by screening 66 patients who filled Bethesda and Amsterdam criteria for LS from health institutions of Brazil. The mean depth of coverage for MSH2, MSH6, MLH1 and PMS2 genes was 7.988, 36.313, 11.899 and 4.772 times, respectively. Ninety-four variants were found in exons and flanking intron/exon regions for the four MMR genes. Twenty-five were pathogenic or VUS and found in 32 patients (7 in MSH2, 5 in MSH6, 12 in MLH1 e 1 in PMS2). All variants were confirmed by Sanger sequencing. The strategy was efficient to reduce time consuming and costs to identify genetic changes at these MMR genes, reducing in three times the number of PCR reactions performed per patient and was efficient in identifying variants at PMS2 gene.

Similar content being viewed by others
References
Giardiello FM, Allen JI, Axilbund JE et al (2014) Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US multi-society task force on colorectal cancer. Gastroenterology 147(2):502–526. doi:10.1053/j.gastro.2014.04.001
Jasperson KW, Tuohy TM, Neklason DW, Burt RW (2010) Hereditary and familial colon. Cancer Gastroenterol 138(6):2044–2058. doi:10.1053/j.gastro.2010.01.054.Hereditary
Hampel H (2016) Genetic counseling and cascade genetic testing in Lynch syndrome. Fam Cancer 15(3):423–427. doi:10.1007/s10689-016-9893-5
Plazzer JP, Sijmons RH, Woods MO et al (2013) The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer 12(2):175–180. doi:10.1007/s10689-013-9616-0
Noll A, Parekh P, Karlitz J (2016) Diagnosis of Lynch syndrome before colorectal resection: does it matter?. Tech Coloproctol 20(4):203–205. doi:10.1007/s10151-016-1433-7
Fishel R, Lescoe MK, Rao MRS et al (1993) The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75(5):1027–1038. doi:10.1016/0092-8674(93)90546-3
Miyaki M, Konishi M, Tanaka K et al (1997) Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet 17(3):271–272. doi:10.1038/ng1197-271
Bronner CE, Baker SM, Morrison PT et al (1994) Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368(6468):258–261. doi:10.1038/368258a0
Nicolaides NC, Papadopoulos N, Liu B et al (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371(6492):75–80. doi:10.1038/371075a0
Van der Klift HM, Mensenkamp AR, Drost M et al (2016) comprehensive mutation analysis of PMS2 in a large cohort of probands suspected of Lynch syndrome or constitutional mismatch repair deficiency syndrome. Hum Mutat 37(11):1162–1179. doi:10.1002/humu.23052
Vasen HF, Blanco I, Aktan-Collan K et al (2013) Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 62(6):812–823. doi:10.1136/gutjnl-2012-304356
Susswein LR, Marshall ML, Nusbaum R et al (2015) Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 18:1–10. doi:10.1038/gim.2015.166
Hoppman-Chaney N, Peterson LM, Klee EW, Middha S, Courteau LK, Ferber MJ (2010) Evaluation of oligonucleotide sequence capture arrays and comparison of next-generation sequencing platforms for use in molecular diagnostics. Clin Chem 56(8):1297–1306. doi:10.1373/clinchem.2010.145441
Talseth-Palmer BA, Bauer DC, Sjursen W et al (2016) Targeted next-generation sequencing of 22 mismatch repair genes identifies Lynch syndrome families. Cancer Med 5(5):929–941. doi:10.1002/cam4.628
Jansen AML, Geilenkirchen MA, Van Wezel T, Jagmohan-Changur SC et al (2016) Whole gene capture analysis of 15 CRC susceptibility genes in suspected Lynch syndrome patients. PLoS ONE 11(6):1–15. doi:10.1371/journal.pone.0157381
Hansen MF, Neckmann U, Lavik LAS et al (2014) A massive parallel sequencing workflow for diagnostic genetic testing of mismatch repair genes. Mol Genet genomic Med 2(2):186–200. doi:10.1002/mgg3.62
Carneiro da Silva F, de Oliveira Ferreira JR, Torrezan GT et al (2015) Clinical and molecular characterization of Brazilian patients suspected to have Lynch syndrome. PLoS One 10(10):e0139753. doi:10.1371/journal.pone.0139753
De Vos M, Hayward BE, Picton S, Sheridan E, Bonthron DT (2004) Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome. Am J Hum Genet 74:954–964
Hayward BE, De Vos M, Valleley EM et al (2007) Extensive gene conversion at the PMS2 DNA mismatch repair locus. Hum Mutat 28(5):424–430. doi:10.1002/humu.20457
Li J, Dai H, Feng Y et al (2015) A comprehensive strategy for accurate mutation detection of the highly homologous PMS2. J Mol Diagnostics 17(5):545–553. doi:10.1016/j.jmoldx.2015.04.001
Zumstein V, Vinzens F, Zettl A, Heinimann K, Koeberle D, Bolli M (2016) Systematic immunohistochemical screening for Lynch syndrome in colorectal cancer : a single centre experience of 486 patients. Swiss Med Wkly 146:1–6. doi:10.4414/smw.2016.14315
Ricciardiello L, Ahnen DJ, Lynch PM (2016) Chemoprevention of hereditary colon cancers: time for new strategies. Nat Rev Gastroenterol Hepatol 13(6):352–361. doi:10.1038/nrgastro.2016.56
Beck NE, Tomlinson IPM, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF (1997) Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal even if the Amsterdam criteria are not fulfilled. Br J Surg 84(2):233–237. doi:10.1002/bjs.1800840228
Zahary MN, Kaur G, Hassan MRA, Singh H, Naik VR, Ankathil R (2012) Germline mutation analysis of MLH1 and MSH2 in Malaysian Lynch syndrome patients. World J Gastroenterol 18(8):814–820. doi:10.3748/wjg.v18.i8.814
Chadwick RB, Pyatt RE, Niemann TH et al (2001) Hereditary and somatic DNA mismatch repair gene mutations in sporadic endometrial carcinoma. J Med Genet 38(7):461–466
Mensenkamp AR, Vogelaar IP, Van Zelst-Stams WAG et al (2014) Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 146(3):643–646.e8. doi:10.1053/j.gastro.2013.12.002
Vaughn CP, Robles J, Swensen JJ et al (2010) Clinical analysis of PMS2: mutation detection and avoidance of pseudogenes. Hum Mutat 31(5):588–593. doi:10.1002/humu.21230
De Leeneer K, De Schrijver J, Clement L et al (2011) Practical tools to implement massive parallel pyrosequencing of PCR products in next generation molecular diagnostics. Plos One 6(9):e25531. doi:10.1371/journal.pone.0025531
Ring KL, Bruegl AS, Allen BA et al (2016) Germline multi-gene hereditary cancer panel testing in an unselected endometrial cancer cohort. Modern Pathololy 29(11):1381–1389. doi:10.1038/modpathol.2016.135
Sie AS, Prins JB, van Zelst-Stams WAG, Veltman JA, Feenstra I, Hoogerbrugge N (2015) Patient experiences with gene panels based on exome sequencing in clinical diagnostics: high acceptance and low distress. Clin Genet 87(4):319–326. doi:10.1111/cge.12433
Pritchard CC, Smith C, Salipante SJ et al (2012) ColoSeq provides comprehensive Lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn 14(4):357–366. doi:10.1016/j.jmoldx.2012.03.002
Hitch K, Joseph G, Guiltinan J, Kianmahd J, Youngblom J, Blanco A (2014) Lynch syndrome patients’ views of and preferences for return of results following whole exome sequencing. J Genet Couns 23(4):539–551. doi:10.1007/s10897-014-9687-6
Tutlewska K, Lubinski J, Kurzawski G (2013) Germline deletions in the EPCAM gene as a cause of Lynch syndrome—literature review. Hered Cancer Clin Pract 11(1):1–9. doi:10.1186/1897-4287-11-9
Yao R, Goetzinger KR (2016) Genetic carrier screening in the twenty-first century. Clin Lab Med 36(2):277–288. doi:10.1016/j.cll.2016.01.003
De Leeneer K, Hellemans J, De Schrijver J, Baetens M, Poppe B, Van Criekinge W, De Paepe A, Coucke P, Claes K (2011) Massive parallel amplicon sequencing of the breast cancer genes BRCA1 and BRCA2: opportunities, challenges, and limitations. Hum Mutat 32(3):335–344. doi:10.1002/humu.21428
De Carvalho MCDCG, Da Silva DCG (2010) Next generation DNA sequencing and its applications in plant genomics. Ciência Rural 40(3):735–744. doi:10.1590/S0103-84782010000300040
Acknowledgements
This work was supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ-Brazil) (Grant No. E26/170.026/2008), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq-Brazil) (Grant Nos. 573806/2008-0, 305873/2014-8), Instituto Nacional para Controle do Cancer (http://www.inctcancer-control.com.br) and Brazilian Ministry of Heath.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in the present work was approved by the Ethics Committee of each institution (CAAE-0254.1.001.007-11) in accordance with the ethical standards and with the Helsinki declaration.
Informed consent
Informed consent was obtained from all patients included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Soares, B.L., Brant, A.C., Gomes, R. et al. Screening for germline mutations in mismatch repair genes in patients with Lynch syndrome by next generation sequencing. Familial Cancer 17, 387–394 (2018). https://doi.org/10.1007/s10689-017-0043-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-017-0043-5