Abstract
Telomeres, DNA structures located at the end of eukaryotic chromosomes, shorten with each cellular cycle. The shortening rate is affected by factors associated with stress, and, thus telomere length has been used as a biomarker of ageing, disease, and different life history trade-offs. Telomere research has received much attention in the last decades, however there is still a wide variety of factors that may affect telomere measurements and to date no study has thoroughly evaluated the possible long-term effect of a storage medium on telomere measurements. In this study we evaluated the long-term effects of ethanol on relative telomere length (RTL) measured by qPCR, using blood samples of magpies collected over twelve years and stored in absolute ethanol at room temperature. We firstly tested whether storage time had an effect on RTL and secondly we modelled the effect of time of storage (from 1 to 12 years) in differences in RTL from DNA extracted twice in consecutive years from the same blood sample. We also tested whether individual amplification efficiencies were influenced by storage time, and whether this could affect our results. Our study provides evidence of an effect of storage time on telomere length measurements. Importantly, this effect shows a pattern of decreasing loss of telomere sequence with storage time that stops after approximate 4 years of storage, which suggests that telomeres may degrade in blood samples stored in ethanol. Our method to quantify the effect of storage time could be used to evaluate other storage buffers and methods. Our results highlight the need to evaluate the long-term effects of storage on telomere measurements, particularly in long-term studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Telomeres are non-coding repetitive and conserved DNA sequences located at the ends of eukaryotic chromosomes that maintain their integrity (Blackburn 1991). Telomeres in vivo are compacted in a chromatin structure that interacts with modified histones, and their ends are associated with other proteins forming a complex denominated shelterin (De Lange 2005). This complex forms a loop (T-loop; Griffith et al. 1999) that protects the single-stranded 3’ overhang end of the chromosome from the DNA damage response machinery that, otherwise, would recognize it as broken DNA and trigger cell replicative senescence or its apoptosis (De Lange 2010).
During the replication process, the DNA polymerase cannot replicate the end of the lagging DNA strand (“the end-replication problem”) and telomeres shorten with each cellular cycle (Blackburn 1991). When telomeres shorten to a critical length, the cell enters a state of replicative senescence or dies via apoptosis (Hemann et al. 2001; Zou et al. 2004). Telomeres then also protect the coding DNA sequence from loss during replication and help to maintain the proliferative capacity of cells (Blackburn 2000).
Moreover, telomeres constitute fragile sites that are more susceptible to oxidative damage than other parts of the genome and are repaired less efficiently (Petersen et al. 1998; Rhee et al. 2011; Coluzzi et al. 2014), so that situations that may cause oxidative stress, such as infection or reproduction (Chatelain et al. 2020), may contribute to telomere loss. This loss can result in an accumulation of defects that may affect the function of the different tissues leading to organismal ageing and disease; (López-Otín et al. 2013; Blackburn et al. 2015) but may also link lifespan and lifestyle (Monaghan and Haussmann 2006) and help to understand how individuals deal with stressful circumstances (Haussmann and Marchetto 2010; Monaghan 2014). For these reasons, telomere biology has become an important topic in different areas of research, such as epidemiology or evolutionary ecology.
The study of telomeres in natural populations has received considerable attention in the last two decades (Monaghan et al. 2018). Telomeres may play an important role in mediating different life-history trade-offs (Young 2018), and telomere length have been shown to be correlated with, for example, survival (Wilbourn et al. 2018) or reproductive success (Eastwood et al. 2019) in a variety of taxa. Besides, the rate at which telomeres shorten is also related to growth (Monaghan and Ozanne 2018), the costs of reproduction (Sudyka et al. 2019), environmental conditions (Spurgin et al. 2018; Foley et al. 2020) or lifespan (Whittemore et al. 2019), and thus, both telomere length and its dynamics may give us insights into the life history strategies of individuals.
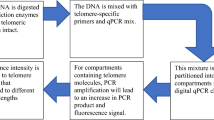
There are different methods available to estimate telomere length (Nakagawa et al. 2004; Nussey et al. 2014), although telomere measurement using real-time quantitative PCR (qPCR; Cawthon 2002, 2009) has probably become the most widespread method in all areas of telomere research (Pepper et al. 2018; Morinha et al. 2020a). qPCR is an inexpensive and fast method that only requires a small amount of DNA (Cawthon 2002) and estimates a relative telomere length (RTL) as the ratio between the amount of telomeric sequence (T) and the amount of sequence of a single-copy reference gene (S) amplified from the same sample. However, several factors can impact telomere measurement by qPCR (reviewed in Morinha et al. 2020a) and there is still heterogeneity regarding sample collection, storage and DNA extraction methods. This is especially important in the case of long-term studies, where samples taken at some point can be analysed quite a long time after collection; in some instances, samples may have not been intended for measuring telomeres.
Different studies have evaluated the impact of different storage methods on telomere measurements. For example, Reichert et al. (2017) found differences in telomere length estimations depending on the storage method in short-term and long-term stored samples, but the effect of storage could be confounded by different extraction methods, which have already been shown to impact telomere measurements by qPCR (Cunningham et al. 2013; Tolios et al. 2015; Raschenberger et al. 2016; Eastwood et al. 2018).
Ethanol is widely used for the long-term storage of blood samples (Kilpatrick 2002; Camacho-Sanchez et al. 2013), since it is easy to obtain, found in most labs, inexpensive and maintains DNA quality for a long period at room temperature. It has also been shown to be a good blood preservation method for telomere measurement by qPCR compared to other storage buffers (Eastwood et al. 2018) and that it yields a high correlation between TL measurements by qPCR and terminal restriction fragment assay (correlation coefficients r ranged from 0.68 to 0.74). However, the suitability of ethanol as a long-term storage medium for samples used in telomere measurements has not previously been tested.
Here, we present a test of the effect of long-term preservation of blood samples in absolute ethanol at room temperature on telomere length measurement by qPCR. We aim, (i) to test if the number of years that samples had been stored in ethanol affects telomere length measurements, using samples stored between 1 and 12 years. Secondly, (ii) we aim to model how the differences in the estimation of RTL from DNA templates extracted from the same blood sample in two consecutive years depend on the duration of storage (1 to 12 years).
Methods
Sample collection and study design
All the samples were collected from a population of Eurasian magpies (Pica pica) located in La Calahorra, Granada, South-eastern Spain (37°10′N, 3°03′W). This population has been monitored since 2005, and blood samples (about 100 µl) from adults and chicks have been collected during each breeding season (March - July) and kept in one millilitre of absolute ethanol (Analytical Reagent Grade) at room temperature for future analysis.
Blood samples were taken from 143 magpies, 118 corresponded to chicks that were sampled soon before fledging, during each breeding season from 2005 to 2017 (age class = 0); and 25 samples corresponded to adult magpies (age class = 1), that were captured during breeding in the same period. Genomic DNA was extracted from the erythrocytes of the whole blood samples by the ammonium acetate precipitation method following exactly the same protocol for all samples (Richardson et al. 2002) between October and November of 2016, 2017 or 2018 and stored at -20ºC. Relative telomere length (RTL) of these samples was estimated by real-time PCR method (see below).
To test whether long-term storage in ethanol could have an effect on the estimates of telomere length we used 91 blood samples that had been stored in absolute ethanol up to eleven years in 2016 (Fig. 1). We extracted DNA from the blood samples and measured their RTL in 2016, and, one year later (2017), we then performed a second DNA extraction in 2017 from the same samples and used this DNA to measure their RTL again. We ensured that ethanol volume was at least ten times larger than sample volume in all cases (Barrett et al. 2012). We could then check whether there were differences in RTL of samples stored in ethanol between consecutive years and whether these differences were related to the time that samples had been stored in ethanol before the second DNA extraction (1 to 12 years). We used between 2 and 23 blood samples taken per year between 2005 and 2016 (mean (SD): 6.4 samples per year (3.7)). Unfortunately, we were unable to perform the second DNA extraction from the samples obtained in 2013 and 2012, so we did not have data of samples stored for 4 and 5 years.
Telomere length measurements by qPCR
All laboratory analyses were carried out at the NERC Biomolecular Analysis Facility of the University of Sheffield (United Kingdom).
DNA sample concentrations were measured using a NanoDrop 8000 Spectrophotometer (ThermoScientific™, Waltham, USA); samples were then diluted to 25 nanograms/microlitre. DNA quality was assessed by agarose gel electrophoresis in a subset of the oldest samples. We additionally performed a high sensitivity D1000 ScreenTape electrophoresis assay with an Agilent 4200 TapeStation system (Agilent Technologies, Santa Clara, USA) in genomic DNA from 4 samples that were extracted twice (in two consecutive years) to assure the integrity of the DNA (see Figs S2, Supplementary material).
Telomere length was assessed by real-time PCR (qPCR) in a monochrome multiplex reaction (Cawthon 2009) with some modifications. We used telc and telg primers for the telomere sequence (Cawthon 2009) and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the reference gene. For this gene, we designed a pair of specific primers for magpies from the GenBank Pica pica Sequence: EF052752. This primer set was clamped with two guanine-cytosine tails on their 5’ to elevate their melting temperature (Tm), to allow us to separate the amplification of the telomere sequence and the GAPDH sequence within the same reaction (see Cawthon 2009 for further explanations). The size of the product amplified with this primer set was 97 bp, similar to the fixed length telomere product. The sequence of the forward and reverse primers for GAPDH including the clamp are 5’- CGGCGGCGGGCGGCGCGGGCTGGGCGGAGCC-AAAGTGGCTCCAATCCCACT-3’ and 5’-GCCCGGCCCGCCGCGCCCGTCCCGCCG-CTGAACTCCCATCCACCCT-3’, respectively. Primer concentration was 900 nM for both pairs of primers and DNA concentration was 25 nanograms per reaction. Reaction volume was 20 µl containing 10 µl of SYBR ™ Select Master Mix (Applied Biosystems™, Warrington, UK).
Each sample was run in duplicate, loaded manually in MicroAmp Fast Optical 96 Well Plates and sealed with MicroAmp Optical Adhesive Film (Applied Biosystems™, Warrington, UK). Samples were randomly distributed on 29 different plates (along with samples belonging to a different study) to avoid bias regarding the year of sampling. All qPCR assays were performed in a QuantStudio 12 K Flex qPCR instrument (Applied Biosystems™, Warrington, UK) and the thermal cycling profile was as follows: Hold Stage: 2 min at 50ºC, 2 min at 95ºC; Stage 1: 2 cycles of 15 s at 94ºC, 10 s at 49ºC; Stage 2: 40 cycles of 15 s at 94ºC, 10 s of 62ºC, 15 s at 74ºC with signal acquisition (for the telomere amplification), 10 s at 84ºC, 15 s at 86 ºC with signal acquisition (for the GAPDH amplification); Melt curve stage: 15 s at 95ºC, 1 min at 60ºC, 15s at 95ªC. The melting curve allowed us to confirm the presence of both amplicons and to evaluate the specificity of the primers (Morinha et al. 2020b).
Each plate included two no-template controls (NTC) and serial dilutions by duplicate (80, 20, 5, 1.25, 0.3125 nanograms/microlitre) of DNA from the same sample. The 20 nanograms/microliter sample of these serial dilutions constituted the calibrator sample, and was used to calculate inter-plate variation and normalized all T/S ratios. We used the software LinRegPCR (Ruijter et al. 2009) to subtract baseline fluorescence and to calculate individual efficiencies and Cq values (cycle at which amplification crosses a fixed threshold) for each well and amplicon (for the telomere sequence and GAPDH amplifications). T/S ratios were calculated following the method described by (Pfaffl 2001):
Where E is the average efficiency of all the reactions in a plate (for telomere sequence and GAPDH amplifications, respectively), and ∆Cq is the difference between the average Cq value of the calibrator sample and the Cq value of the sample being analysed (for each sequence, respectively).
Amplification efficiencies were on average 1.94 (0.04) for telomeres and 1.90 (0.03) for GAPDH. The average intra-plate standard deviations of Cq values were on average 0.079 (0.017) for telomeres and 0.078 (0.018) for GAPDH. Intra-plate and inter-plate T/S ratio standard deviations were on average 0.036 (0.007) and 0.063 (0.033), respectively. R² for the log-linear phase of the amplification curve were on average 0.99 (0.0003) for telomeres and 0.99 (0.003) for GAPDH. Samples whose standard deviation of T/S ratios between replicates was over 0.05 were excluded from further analysis.
T/S ratios varied between 0.21 and 2.14. All T/S ratios were normalized to account for inter-run variability by dividing the average T/S ratio of the duplicates of each sample by the average T/S ratio of the calibrator sample of the plate where the sample was run. This normalized value is our estimate of relative telomere length (RTL).
Statistical analyses
All statistical analyses were performed in R version 4.2.0 ( R Core Team 2020). All continuous variables were z-standardized and categorical variables were mean-centered (Schielzeth 2010).
Sample storage duration and RTL
To explore whether the time (years) that samples had been preserved in ethanol affected the estimation of RTL, we constructed a linear mixed model (LMM, package lme4 version 1.1–29; Bates et al. 2015) in which RTL was the dependent variable and the number of years of storage (storage time) and the age class of individuals when sampled (chick: 0, adult: 1) were also included as factors. Visual inspection of the raw data suggested that the relationship between the difference in RTL and storage time (number of years elapsed between blood sampling and the second DNA extraction) could be polynomial, so we tested whether including a quadratic and a cubic term of the storage time significantly improved the model fit. The linear model significantly differed from the quadratic model (χ2 = 50.1, p-value < 0.001) and the quadratic model significantly differed from the best fitting cubic model (χ2 = 33.7, p-value < 0.001; see Table S1, Supplementary material for a comparison of the three models), so we finally included storage time and its orthogonal polynomials of degree two and three as continuous fixed effects. We also included as random factors individual identity to account for the samples that had been extracted twice and nest identity because there were siblings in our data set. The analysis initially included 234 DNA samples extracted from 143 blood samples (individuals) belonging to 117 nests, but 5 samples were excluded because they didn’t meet our inclusion criteria, so the final sample size used for the analysis was 229 samples extracted from 139 individuals belonging to 114 nests.
We additionally used 91 samples that had been extracted twice in two consecutive years and constructed a linear mixed model (LMM, package lme4 version 1.1–29; Bates et al. 2015) in which RTL of the second extraction was the dependent variable and the RTL of the first extraction, the number of years that the sampled had been stored before the second extraction (storage time) and the two-way interaction between them were included as fixed effects; individual age class when sampling (chick: 0, adult: 1) was included as a factor. Age class when sampling was not significant (β = 0.01; 95% CI = -0.07, 0.08; t = 0.41; p = 0.69) and explained little variance, so it was not included in the final model. Nest identity was included as a random effect. Three samples initially included in this analysis did not meet our inclusion criteria, so were excluded alongside their paired sample.
We also tested whether RTL in the first extraction was greater than RTL in the second extraction using a one-tailed paired T-test.
Modelling the effect of storage time on RTL estimation
We also used 88 blood samples (those extracted twice in two consecutive years) to model how RTL could vary depending on the duration of storage (1 to 12 years). To this aim, we constructed a linear model (LM) in which the difference in RTLs estimated using two DNA samples extracted in two consecutive years from the same blood sample was the dependent variable, and we included storage time and its orthogonal polynomials of degree two and three as continuous fixed effects, although the last one was dropped from the model because it was not significant (β = -0.0001; 95% CI = -0.07, 0.08; t = -0.38; p = 0.70) and explained little variance. The first estimate of RTL was also included as a continuous fixed effect. This model would allow us to estimate for each sample the relative amount of telomere sequence lost (differences in RTLs) depending on the time of storage and determine any temporal pattern.
The difference in RTLs and the first estimated RTL are not statistically independent, and this dependency can lead to a biased estimate of the relationship between these two variables caused by a statistical artefact called “regression to the mean” (Barnett et al. 2005). For this reason we additionally constructed a LM in which the differences in RTL were corrected for this effect using the equation described in Verhulst et al. (2013) so that a corrected value for the differences (D) was the dependent variable and the first estimate of RTL was included as continuous fixed effect.
Individual amplification efficiencies
We tested whether storage time could have an effect on individual amplification efficiencies and their differences by constructing three linear mixed models (LMMs, package lme4 version 1.1–29; Bates et al. 2015) with all the RTLs used in this study (N = 229) in which alternatively, individual efficiency of the telomere amplification, individual efficiency of the GAPDH amplification and the differences between both efficiencies were the dependent variables, with storage time included as a fixed effect and plate identity included as a random factor. Additionally, we tested whether differences in individual efficiencies could affect the estimation of differences between RTLs of the same sample extracted twice in two consecutive years by constructing a linear model in which this difference was the dependent variable and storage time, the differences in efficiencies between telomere and GAPDH amplification from the first extraction, the differences in efficiencies between telomere and GAPDH amplification from the second extraction and two two-way interactions between storage time and the differences in efficiencies from both extractions were included as fixed effects.
Well position effects
We used a subset of 9 plates to ascertain whether our RTL estimates could be affected by well position effects, this is, variations in RTLs due to variations in temperature across the qPCR instrument heating block. We then constructed a linear model in which individual well T/S ratio was the dependent variable and the well closest distance to the x-axis edge of the plate (from 0 to 3) and the well closest distance to the y-axis edge of the plate (from 0 to 5) were included as fixed effects. Standard samples and NTC were always placed in the same well positions and thus were excluded from this analysis which finally included 756 T/S ratios.
Results
Sample storage duration and RTL
There was a significant cubic relationship between the number of years a sample had been stored and the RTL estimated from it (Table 1), and samples that had been preserved for more years tended to have shorter RTL (Figure S1, Supplementary material). On the other hand, there were no significant differences in RTL between samples from fledglings and adults in our data set (Table 1).
RTLs obtained from the second extraction of samples extracted twice in consecutive years differed from RTLs of first extraction depending of the storage time (Table 2; Fig. 2).
Indeed, RTLs obtained from the second extraction were significantly smaller than RTLs from the first extraction (one-tailed T-test: t = -3.62, df = 87, p < 0.001), and this difference was driven by RTLs of the second extraction being smaller for the samples that had been stored for 1 to 3 years (one-tailed T-test: t = -9.07, df = 25, p < 0.001), but not for the samples that had been stored for a longer period of time (one-tailed T-test: t = -2.82, df = 61, p = 0.99).
Modelling storage-time effects on RTL
The differences in RTL decreased for samples stored between one and four years until they drop to approximately zero (Table 3; Fig. 3). This suggests that there is degradation of telomeres causing shortening during the first four years of storage in ethanol and then the degradation stops. Our results also suggest that this degradation is dependent on telomere length, as the first RTL is positively and significantly related to the differences in RTL (Table 3) and this relationship is not a result of a regression to the mean effect since the corrected value (D) was significantly explained by RTL1 (see Supplementary material Table S2).
Individual amplification efficiencies
Individual amplification efficiencies were negatively affected by storage time for both telomere and GAPDH sequences, however the difference between both efficiencies was not (Table S3). Indeed, the differences in efficiencies between both sequences from the first and the second extractions in relation to storage time did not seem to affect the differences in RTLs of the same sample extracted twice in two consecutive years (Table S4).
Well position effects
Individual well T/S ratios were not related to the well closest distance neither to x-axis (β = 0.001, CI = -0.02, 0.03; t = 0.12; p = 0.90) nor to y-axis edge (β = 0.006, CI = -0.01, 0.02; t = 0.75; p = 0.45).
Discussion
Our study presents the first comprehensive evaluation of the long-term effect of a storage medium on the estimation of relative telomere length. Different studies have shown that different storage media or conditions may affect T/S estimates, however, most of them have been carried out in the short-term. Only one study analysed RTLs of bird samples taken longitudinally over 14 years and stored in different ways, finding evidence of degradation/fragmentation in DNA extracted from blood samples stored in dried blood spot cards, resulting in shorter RTLs (Reichert et al. 2017) compared to samples stored as frozen whole blood or frozen extracted DNA, and also to samples that had been stored dried blood spot cards for two months. On the contrary, a different study found that samples stored as dried blood spot cards yielded higher RTLs compared to frozen whole blood, but in a shorter storage time (9 days; Zanet et al. (2013)). Other studies have evaluated storage media and/or storage conditions such as temperature on telomere measurements (Dagnall et al. 2017; Tolios et al. 2015; Eastwood et al. 2018), finding differences in RTLs depending on the storage medium and not on the temperature; again, the storage time was short (up to 7 months) in those cases. Some studies have used ethanol to preserve samples in the long term and one of them has reported no storage time effect of ethanol at room temperature in RTL of a subset of samples (Spurgin et al. 2018b), but the authors do not provide information about how long samples were stored before DNA extraction.
Our first cross-sectional analysis showed that there is a negative cubic effect of the time samples are stored in ethanol at room temperature on their relative telomere length estimates (Table 1). Although we cannot discard that telomere length in our study population has changed over the study period, maybe because some unknown environmental factor affecting telomere length has changed over the years, causing an increase in telomere length, we think this is an unlikely explanation. First, our population has not shown any other linear trend in other reproductive variables during the study period. Second, the samples used in this study are a mix of individuals of many different ages, which makes very difficult to record a hypothetical positive trend in telomere length over the years; because many of them were not sampled in the year they were born, for a particular year samples used consist of some individuals born that year and many others of different ages. This data structure would not recover a temporal change in telomere length related to some environmental variable. In the same way, we think we can discard the possibility that we could have randomly selected individuals with longer telomere length in recent years, which would lead to the same pattern, because the methods used to capture and sample individuals (either adults or chicks) have not changed in recent years.
In any case, our second analysis showed that the second RTL of samples extracted twice in consecutive years is smaller than the first RTL only for those samples that had been stored between one and three years (Table 2; Fig. 2). Indeed, when we model how the storage time affects the differences in RTL between those paired samples (Table 3) we observe that telomeres suffer from degradation during the first 3 or 4 years of storage, and after this time further degradation appears to halt (Fig. 3).
The loss of telomere sequence between consecutive DNA extractions is not likely to be due to genomic DNA degradation, since in that case, samples stored for larger periods of time should experience higher levels of telomere sequence loss; however, our data show the opposite pattern: samples stored for fewer years before the first RTL estimation lost more telomere sequence between two consecutive DNA extractions and RTL estimations. Indeed, high sensitivity D1000 ScreenTape electrophoresis didn’t show degradation in samples that were stored one year before the second extraction (Supplementary material, Fig. S2). Also, although individual efficiencies may be affected by storage time, this effect is similar for both telomere and GAPDH sequences and relatively small (Table S3). Only a differential change in individual efficiencies between telomere and GAPDH amplifications over the time could have yielded the pattern of telomere loss found in our study, and this seems not to be the case (Table S4).
One possible explanation for this phenomenon could be related to telomere composition and structure itself: telomere repeats are rich in guanine, which is the base most susceptible to oxidation (Wang et al. 2010), and they constitute fragile sites that are susceptible to accumulate oxidative damage (Petersen et al. 1998). This damage can be caused by exogenous sources and/or cell metabolism (De Bont and van Larebeke 2004). Moreover, telomere damage is repaired less frequently than other parts of the genome (Coluzzi et al. 2014; Rhee et al. 2011) and the 3’ overhang lacks a complementary template to be repaired from (Ahmed and Lingner 2018). In vivo, telomeres are usually protected by the shelterin complex until damage can be repaired, and this complex also regulates their repair (De Lange 2005). However, when cells (erythrocytes in this case) are placed in ethanol, the shelterin proteins lose their tertiary structure and precipitate and t-loops are disarranged, leaving the telomere sequence and its fragile sites exposed. It could be also possible that, due to the polar nature of its hydroxyl group, ethanol could dissolve reactive oxygen species or other oxidizing agents already present in the cell that would continue damaging the exposed telomere sequence until these agents are reduced. This could explain the temporal pattern of telomere degradation that we have found (Fig. 3). Moreover, the size of the t-loop correlates with telomere length (Griffith et al. 1999), which could also explain the length dependent degradation (Table 3). This explanation also suggests that the amount of telomere shortening that a sample suffers could also depend on the redox balance of their cells in the time of sampling, what would explain the percentage of variance not explained by our model (Table 3).
Storage-time effect
Our design allowed us to evaluate the long term effect of storage (up to 12 years) in only two years. This kind of design promises to be a useful tool to evaluate this kind of effects in a short period of time and could be applied to any other storage conditions and methods for telomere measurement. We have also showed that storage in absolute ethanol at room temperature has long-term effects on telomere measurement, contrary to what was suggested by a previous study (Eastwood et al. 2018). In the cited article, authors compared RTLs obtained from the same samples stored in absolute ethanol and other two storage media for 7 months at 4ºC; they also compared those RTLs with absolute measures of telomeres obtained by pulse gel electrophoresis terminal restriction fragment analysis and concluded that ethanol was the best preservative among the 3 tested. Our study differs on the time that samples were kept in ethanol and the temperature at which they were stored, what could be a key factor in the degradation process. In any case, storage effects should be tested when ethanol is used as preservative. Different studies and reviews emphasize the need of keeping a consistent method for sampling and storage as the best strategy (Nussey et al. 2014; Reichert et al. 2017; Morinha et al. 2020a). However, we have shown here that even under constant conditions, loss of telomere sequence may occur and that other factors such as the initial telomere length of the sample when it was collected can have an effect in this loss.
The only way to experimentally determine a correction coefficient would be to obtain RTL measurements from freshly collected samples, and after a few years re-estimate RTL and then validate the correction, but even in this way, differences in procedures between years may have an effect in the correction. Pre-analytical and analytical conditions are considered as an important source of error in RTL measurements by qPCR and could have influenced our results (Morinha et al. 2020a); however, we have tried to reduce this possible effect in different ways: all samples were extracted using exactly the same protocol; we have used multiplex qPCR instead of singleplex PCR, which reduces pipetting mistakes; samples collected in different years were randomly distributed in the plates; we have normalized all T/S ratios to account for inter-run differences and thus between years; we have also tested whether a change in individual efficiencies of both sequences could have yielded the pattern found and we have tested in a subset of samples whether well position effects could have influenced RTL measurements. Moreover, although our experimental design included some gaps, such as lack of data for samples stored for 4 and 5 years and low sample size in samples stored for 2 and 3 years, the explanatory power of the model suggests that the results are consistent (R², Table 3).
Consequences of telomere degradation
Different longitudinal studies have reported that a small percentage of individuals show telomere elongation in tissues in which telomeres are commonly not elongated (Gorbunova and Seluanov 2009). In most cases, this anomaly has been attributed to measurement error (e.g. Salomons et al. 2009; Chen et al. 2011; Steenstrup et al. 2013) and although elongation can be disentangled from measurement error (Simons et al. 2014), this method requires quite large sample sizes and multiple samples from the same individual, which can be difficult to achieve for determined species in wild populations. However, the possible degradation of telomeres during preservation has been rarely considered, though, a length dependent shortening of telomeres during sampling storage along with inconsistencies in the time past between sampling and DNA extraction could be responsible of an apparent elongation of telomeres: if some time pass between the sampling of blood and DNA extraction in a set of individuals, which are later re-sampled and their DNA extracted straight away, we expect telomere degradation in the first set of samples (first telomere length measurement), but not in the second set of samples (non-degraded telomeres), in such a way that the second measurement could produce larger telomeres estimates than the first measurement, in particular in individuals with larger telomeres, because long telomeres would shorten during storage more that short telomeres; this would result in a false elongation effect in the second sampling only apparent in a subset of individuals with longer telomeres. Recently many studies are finding low individual consistency in telomere length across individuals´ life with some individuals showing telomere elongation (Fairlie et al. 2016; Spurgin et al. 2018a), suggesting a complex and dynamic relationship between telomere length, environmental effects and physiology; however, as we have shown here, the possible effect of storage must be carefully evaluated before drawing inferences about the biological meaning of telomere elongation.
On the other hand, the effect of telomere shortening during storage could also be confounded to some extent with cohort effects in longitudinal studies. In this type of studies samples from different years are commonly stored for different periods of time before being extracted and/or analysed, and the possible effect of long-term storage on telomere measurements should be carefully evaluated. Ignoring long-term storage effects can lead to erroneous inferences about changes of telomere length in natural populations with time or about the possible relationship of year related variables, such as environmental variables, with telomere length and telomere dynamics.
Conclusions
Our study provides evidence that storage time affects telomere length measurements when blood samples are stored in ethanol at room temperature. Specifically, this effect shows a temporal pattern of loss of telomere sequence during the first 3 or 4 years of storage, after which the loss halts. We also present a method to quantify the effect of storage time. This approach could be used to evaluate other methods of storage and types of storage media. Our results stress the need to evaluate long-term effects of sample storage on telomere measurements. This is an important consideration in long-term studies where samples were not originally intended for telomere measurements but stored and are used for telomere analysis at a later date.
Data and code availability
Data and code will be available in Dryad repository.
References
Ahmed W, Lingner J (2018) Impact of oxidative stress on telomere biology. Differentiation 99:21–27. https://doi.org/10.1016/j.diff.2017.12.002
Barnett AG, van der Pols JC, Dobson AJ (2005) Regression to the mean: What it is and how to deal with it. Int J Epidemiol 34:215–220. https://doi.org/10.1093/ije/dyh299
Barrett ELB, Boner W, Mulder E et al (2012) Absolute standards as a useful addition to the avian quantitative PCR telomere assay. J Avian Biol 43:571–576. https://doi.org/10.1111/j.1600-048X.2012.05787.x
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Blackburn EH (1991) Structure and function of telomeres. Nature 350:569–573. https://doi.org/10.1038/350569a0
Blackburn EH (2000) Telomere states and cell fates. Nature 408:53–56. https://doi.org/10.1038/35040500
Blackburn EH, Epel ES, Lin J (2015) Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Sci (80-) 350:1193–1198. https://doi.org/10.1126/science.aab3389
Camacho-Sanchez M, Burraco P, Gomez-Mestre I, Leonard JA (2013) Preservation of RNA and DNA from mammal samples under field conditions. Mol Ecol Resour 13:663–673. https://doi.org/10.1111/1755-0998.12108
Cawthon RM (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res 30:1–6. https://doi.org/10.1093/nar/30.10.e47
Cawthon RM (2009) Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 37:e21. https://doi.org/10.1093/nar/gkn1027
Chatelain M, Drobniak SM, Szulkin M (2020) The association between stressors and telomeres in non-human vertebrates: a meta-analysis. Ecol Lett 23:381–398. https://doi.org/10.1111/ele.13426
Chen W, Kimura M, Kim S et al (2011) Longitudinal versus cross-sectional evaluations of leukocyte telomere length dynamics: Age-dependent telomere shortening is the rule. Journals Gerontol - Ser A Biol Sci Med Sci 66 A:312–319. https://doi.org/10.1093/gerona/glq223
Coluzzi E, Colamartino M, Cozzi R et al (2014) Oxidative Stress Induces Persistent Telomeric DNA Damage Responsible for Nuclear Morphology Change in Mammalian Cells. PLoS ONE 9:e110963. https://doi.org/10.1371/journal.pone.0110963
Cunningham JM, Johnson RA, Litzelman K et al (2013) Telomere Length Varies By DNA Extraction Method: Implications for Epidemiologic Research. Cancer Epidemiol Biomarkers Prev 22:2047–2054. https://doi.org/10.1158/1055-9965.EPI-13-0409
Dagnall CL, Hicks B, Teshome K et al (2017) Effect of pre-analytic variables on the reproducibility of qPCR relative telomere length measurement. https://doi.org/10.1371/journal.pone.0184098
De Bont R, van Larebeke N (2004) Endogenous DNA damage in humans: A review of quantitative data. Mutagenesis 19:169–185. https://doi.org/10.1093/mutage/geh025
De Lange T (2010) How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol 75:167–177. https://doi.org/10.1101/sqb.2010.75.017
De Lange T (2005) Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110
Eastwood JR, Hall ML, Teunissen N et al (2019) Early-life telomere length predicts lifespan and lifetime reproductive success in a wild bird. Mol Ecol 28:1127–1137. https://doi.org/10.1111/mec.15002
Eastwood JR, Mulder E, Verhulst S, Peters A (2018) Increasing the accuracy and precision of relative telomere length estimates by RT qPCR. Mol Ecol Resour 18:68–78. https://doi.org/10.1111/1755-0998.12711
Fairlie J, Holland R, Pilkington JG et al (2016) Lifelong leukocyte telomere dynamics and survival in a free-living mammal. Aging Cell 15:140–148. https://doi.org/10.1111/acel.12417
Foley NM, Petit EJ, Brazier T et al (2020) Drivers of longitudinal telomere dynamics in a long-lived bat species, Myotis myotis. Mol Ecol 29:2963–2977. https://doi.org/10.1111/mec.15395
Gorbunova V, Seluanov A (2009) Coevolution of telomerase activity and body mass in mammals: From mice to beavers. Mech Ageing Dev 130:3–9. https://doi.org/10.1016/j.mad.2008.02.008
Griffith JD, Comeau L, Rosenfield S et al (1999) Mammalian telomeres end in a large duplex loop. Cell 97:503–514. https://doi.org/10.1016/S0092-8674(00)80760-6
Haussmann MF, Marchetto NM (2010) Telomeres: Linking stress and survival, ecology and evolution. Curr Zool 56:714–727. https://doi.org/10.1093/czoolo/56.6.714
Hemann MT, Strong MA, Hao LY, Greider CW (2001) The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67–77. https://doi.org/10.1016/S0092-8674(01)00504-9
Kilpatrick CW (2002) Noncryogenic preservation of mammalian tissues for DNA extraction: An assessment of storage methods. Biochem Genet 40:53–62. https://doi.org/10.1023/A:1014541222816
Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmerTest Package: Tests in Linear Mixed Effects Models. J Stat Softw 82:1–26. doi:https://doi.org/10.18637/jss.v082.i1. https://doi.org/10.18637/jss.v082.i13
Lefcheck J (2016) piecewiseSEM: Piecewise structural equation modeling. R package version 1.2.1. Methods Ecol Evol 7:573–579. https://doi.org/10.1111/2041-210X.12512
López-Otín C, Blasco MA, Partridge L et al (2013) The hallmarks of aging. Cell 153:1194–1217
Monaghan P (2014) Organismal stress, telomeres and life histories. J Exp Biol 217:57–66. https://doi.org/10.1242/jeb.090043
Monaghan P, Eisenberg DTA, Harrington L, Nussey D (2018) Understanding diversity in telomere dynamics. Phil Trans R Soc B 373:20160435. https://doi.org/10.1098/RSTB.2016.0435
Monaghan P, Haussmann MF (2006) Do telomere dynamics link lifestyle and lifespan? Trends Ecol Evol 21:47–53. https://doi.org/10.1016/j.tree.2005.11.007
Monaghan P, Ozanne SE (2018) Somatic growth and telomere dynamics in vertebrates: Relationships, mechanisms and consequences. Philos Trans R Soc B Biol Sci 373:20160446. https://doi.org/10.1098/rstb.2016.0446
Morinha F, Magalhães P, Blanco G (2020a) Standard guidelines for the publication of telomere qPCR results in evolutionary ecology. Mol Ecol Resour 20:635–648. https://doi.org/10.1111/1755-0998.13152
Morinha F, Magalhães P, Blanco G (2020b) Different qPCR master mixes influence telomere primer binding within and between bird species. J Avian Biol 51:1–9. https://doi.org/10.1111/jav.02352. https://doi.org/10.1111/jav.02352
Nakagawa S, Gemmell NJ, Burke T (2004) Measuring vertebrate telomeres: Applications and limitations. Mol Ecol 13:2523–2533. https://doi.org/10.1111/j.1365-294X.2004.02291.x
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142. https://doi.org/10.1111/j.2041-210x.2012.00261.x
Nussey DH, Baird D, Barrett E et al (2014) Measuring telomere length and telomere dynamics in evolutionary biology and ecology. Methods Ecol Evol 5:299–310. https://doi.org/10.1111/2041-210X.12161
Pepper GV, Bateson M, Nettle D (2018) Telomeres as integrative markers of exposure to stress and adversity: A systematic review and meta-analysis. R Soc Open Sci 5. https://doi.org/10.1098/rsos.180744
Petersen S, Saretzki G, Zglinicki T, Von Von T (1998) Preferential Accumulation of Single-Stranded Regions in Telomeres of Human Fibroblasts. Exp Cell Res 239:152–160. https://doi.org/10.1006/excr.1997.3893
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:16–21. https://doi.org/10.1093/nar/29.9.e45
R Core Team (2020) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. https://www.r-project.org/. https://www.r-project.org/
Raschenberger J, Lamina C, Haun M et al (2016) Influence of DNA extraction methods on relative telomere length measurements and its impact on epidemiological studies. Sci Rep 6:1–10. https://doi.org/10.1038/srep25398
Reichert S, Froy H, Boner W et al (2017) Telomere length measurement by qPCR in birds is affected by storage method of blood samples. Oecologia 184:341–350. https://doi.org/10.1007/s00442-017-3887-3
Rhee DB, Ghosh A, Lu J et al (2011) Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 10:34–44. https://doi.org/10.1016/j.dnarep.2010.09.008
Richardson DS, Jury FL, Blaakmeer K et al (2002) Parentage assignment and extra-group paternity in a cooperative breeder: the Seychelles warbler (Acrocephalus sechellensis). Mol Ecol 10:2263–2273. https://doi.org/10.1046/j.0962-1083.2001.01355.x
Ruijter JM, Ramakers C, Hoogaars WMHH et al (2009) Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. https://doi.org/10.1093/nar/gkp045
Schielzeth H (2010) Simple means to improve the interpretability of regression coefficients. Methods Ecol Evol 1:103–113
Simons MJP, Stulp G, Nakagawa S (2014) A statistical approach to distinguish telomere elongation from error in longitudinal datasets. Biogerontology 15:99–103. https://doi.org/10.1007/s10522-013-9471-2
Spurgin LG, Bebbington K, Fairfield EA et al (2018a) Spatio-temporal variation in lifelong telomere dynamics in a long‐term ecological study. J Anim Ecol 87:187–198. https://doi.org/10.1111/1365-2656.12741
Spurgin LG, Bebbington K, Fairfield EA et al (2018b) Spatio-temporal variation in lifelong telomere dynamics in a long-term ecological study. J Anim Ecol 87:187–198. https://doi.org/10.1111/1365-2656.12741
Steenstrup T, Hjelmborg JVB, Kark JD et al (2013) The telomere lengthening conundrum - Artifact or biology? Nucleic Acids Res 41:1–7. https://doi.org/10.1093/nar/gkt370
Sudyka J, Arct A, Drobniak SM et al (2019) Birds with high lifetime reproductive success experience increased telomere loss. Biol Lett 15:9–12. https://doi.org/10.1098/rsbl.2018.0637
Tolios A, Teupser D, Holdt LM (2015) Preanalytical conditions and DNA isolation methods affect telomere length quantification in whole blood. PLoS ONE 10:1–14. https://doi.org/10.1371/journal.pone.0143889
Verhulst S, Aviv A, Benetos A et al (2013) Do leukocyte telomere length dynamics depend on baseline telomere length? An analysis that corrects for “regression to the mean”. Eur J Epidemiol 28:859–866. https://doi.org/10.1007/s10654-013-9845-4
Wang Z, Rhee DB, Lu J et al (2010) Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet 6:28. https://doi.org/10.1371/journal.pgen.1000951
Whittemore K, Vera E, Martínez-Nevado E et al (2019) Telomere shortening rate predicts species life span. Proc Natl Acad Sci U S A 116:15122–15127. https://doi.org/10.1073/pnas.1902452116
Wilbourn RV, Moatt JP, Froy H et al (2018) The relationship between telomere length and mortality risk in non-model vertebrate systems: A meta-analysis. Philos Trans R Soc B Biol Sci 373:20160447. https://doi.org/10.1098/rstb.2016.0447
Young AJ (2018) The role of telomeres in the mechanisms and evolution of life-history trade-offs and ageing. Philos Trans R Soc B Biol Sci 373:20160452. https://doi.org/10.1098/rstb.2016.0452
Zanet DAL, Saberi S, Oliveira L et al (2013) Blood and Dried Blood Spot Telomere Length Measurement by qPCR: Assay Considerations. PLoS ONE 8. https://doi.org/10.1371/journal.pone.0057787
Zou Y, Sfeir A, Gryaznov SM et al (2004) Does a sentinel or a subset of short telomeres determine replicative senescence? Mol Biol Cell 15:3709–3718. https://doi.org/10.1091/mbc.e04-03-0207
Acknowledgements
We thank the staff of the Molecular Ecology Laboratory and UK NERC Environmental Omics Facility, especially Natalie dos Remedios for her help during the optimization of the qPCR assay and Mirre Simons for providing the script for the qPCR data analysis and his help with the conceptual aspects of the technique.
Funding
This study was supported by the Spanish Ministry of Science through a project (CGL2014-55362-P), a predoctoral Spanish grant (BES-2015-075675) and a Juan de la Cierva –Incorporación Fellowship (IJC2018-036411-I).
Funding for open access charge: Universidad de Granada / CBUA
Author information
Authors and Affiliations
Contributions
MP conceived the idea, designed the methodology, performed lab work and analyses and wrote the manuscript; MMM and JGM performed field work and supervised data analysis and writing; DAD and TB supervised the lab work and writing. All authors contributed critically to the drafts and gave final approval for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethics approval
The research presented here was conducted in accordance with all applicable laws and rules set forth by the Spanish government and Junta de Andalucía. The required permissions were obtained from the Consejería de Agricultura, Ganadería, Pesca y Desarrollo Sostenible (Delegación Territorial de Desarrollo Sostenible de Granada).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Precioso, M., Molina-Morales, M., Dawson, D.A. et al. Effects of long-term ethanol storage of blood samples on the estimation of telomere length. Evol Ecol 36, 915–931 (2022). https://doi.org/10.1007/s10682-022-10198-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10682-022-10198-1