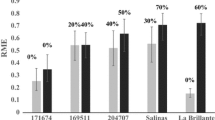
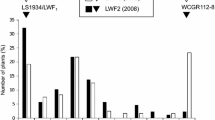
Abstract
Fusarium oxysporum f. sp. lactucae (FOLac) is responsible for significant economic losses across major lettuce-producing regions around the world. Thus far, only FOLac race 1 isolates have been reported associated with Fusarium wilt outbreaks in Brazil. The most sustainable strategy for disease control is the pyramidization of race-specific resistance factors in lettuce cultivars. The loose-leafy cultivar ‘Vanda’ was found as one of the most promising sources of resistance to FOLac race 1. The genetic basis of this resistance was determined by analyzing the reaction to this pathogen of segregating populations derived from the cross ‘Gisele’ (susceptible) × ‘Vanda’ (pollen donor). A single molecular marker-genotyped F1 hybrid plant was selfed and individual plants of a segregating F2 population as well as 63 families F2:F3 were inoculated with a FOLac race 1 isolate by using the root-dipping method (3 × 106 conidia/ml). Our results confirmed the high levels of resistance of ‘Vanda’ even under very harsh experimental conditions. Overall, the reaction of the F1 plants and the segregating patterns of the F2 population (n = 82) and of the F2:F3 families (n = 838 plants) fit a single dominant gene/locus model. However, the phenotypic expression of resistance might suffer effects of additional genetic factor(s) (e.g., locus dosage, minor modifying genes, and incomplete penetrance). Notwithstanding, the high levels of FOLac race 1 resistance and its relatively simple genetic control makes ‘Vanda’ a major germplasm source for lettuce-breeding programs aiming to incorporate this trait in a wide array of elite lines from distinct varietal groups.


Similar content being viewed by others
References
Allex CF (1999) Computational methods for fast and accurate DNA fragment assembly. Ph.D. Thesis. University of Wisconsin, Wisconsin, Madison, USA. p 222
Alon H, Katan J, Kedar N (1974) Factors affecting penetrance of resistance to Fusarium oxysporum f. sp. lycopersici in tomato. Phytopathology 64:455–461
Aruga D, Tsuchiya N, Matsumura H, Matsumoto E, Hayashida N (2012) Analysis of RAPD and AFLP markers linked to resistance to Fusarium oxysporum f. sp. lactucae race 2 in lettuce (Lactuca sativa L.). Euphytica 187:1–9
Boiteux LS, Fonseca MEN, Simon PW (1999) Effects of plant tissue and DNA purification method on randomly amplified polymorphic DNA based genetic fingerprinting analyses in carrot. J Am Soc Hortic Sci 124:32–38
Cabral CS, Reis A (2013) Screening of lettuce accessions for resistance to Fusarium oxysporum f. sp. lactucae race 1. Trop Plant Pathol 38:272–281
Cabral CS, Brunelli KR, Costa H, Fonseca MEN, Boiteux LS, Reis A (2014) Identification of Fusarium oxysporum f. sp. lactucae race 1 as the causal agent of lettuce wilt in Brazil. Trop Plant Pathol 39:197–202
Cabral CS, Fonseca MEN, Brunelli KR, Rossato M, Costa H, Boiteux LS, Reis A (2018) Relationships among Brazilian and worldwide isolates of Fusarium oxysporum f. sp. lactucae race 1 inferred from ribosomal intergenic spacer (IGS-rDNA) region and EF-1α gene sequences. Eur J Plant Pathol 152:81–94
D’Andrea L, Felber F, Guadagnuolo R (2008) Hybridization rates between lettuce (Lactuca sativa) and its wild relative (L. serriola) under field conditions. Environ Biosaf Res 7:61–71
Davis RM, Colyer PD, Rothrock CS, Kochman JR (2006) Fusarium wilt of cotton: population diversity and implications for management. Plant Dis 90:692–703
Dziechciarkova M, Lebeda A, Dolezalova I, Astley D (2004) Characterization of Lactuca spp. germplasm by protein and molecular markers—a review. Plant Soil Environ 50:47–58
Fujinaga M, Ogiso H, Tsuchiya N, Saito H (2001) Physiological specialization of Fusarium oxysporum oxysporum f. sp. lactucae, a causal organism of fusarium root rot of crisp head lettuce. J Gen Plant Pathol 67:205–206
Fujinaga M, Ogiso H, Tsuchiya N, Saito H (2003) Race 3, a new race of Fusarium oxysporum f. sp. lactucae determined by a differential system with commercial cultivars. J Gen Plant Pathol 69:23–28
Gilardi G, Franco Ortega S, van Rijswick PCJ, Ortu G, Gullino ML, Garibaldi A (2016) A new race of Fusarium oxysporum f. sp. lactucae of lettuce. Plant Pathol 66:677–688
Hu J, Ochoa OE, Truco MJ, Vick BA (2005) Application of the TRAP technique to lettuce (Lactuca sativa L.) genotyping. Euphytica 144:225–235
Huang JH, Lo CT (1998) Wilt of lettuce caused by Fusarium oxysporum in Taiwan. Plant Pathol Bull 7:150–153
Hubbard JC, Gerik JS (1993) A new disease of lettuce incited by Fusarium oxysporum f. sp. lactucum forma specialis nov. Plant Dis 77:750–754
Hwang CF, Bhakta AV, Truesdell GM, Pudlo WM, Williamson VM (2000) Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12:1319–1329
Kesseli RV, Michelmore RW (1986) Genetic variation and phylogenies detected from isozyme markers in species of Lactuca. J Hered 77:324–331
Kesseli R, Ochoa O, Michelmore RW (1991) Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce (L. sativa). Genome 34:430–436
Lopes CA, Quezado-Duval A, Reis A (2010) Doenças da Alface. Brasília: Embrapa Vegetable Crops, Brasília, DF, Brazil
Matheron ME, Porchas M (2010) Evaluation of soil solarization and flooding as management tools for Fusarium wilt of lettuce. Plant Dis 94:1323–1328
Matuo T, Motohashi S (1967) On Fusarium oxysporum f. sp. lactucae n.f. causing root of lettuce. Trans Mycol Soc Jpn 8:13–15
McCreight JD, Matheron ME, Tickes BR, Plantts B (2005) Fusarium wilt race 1 on lettuce. HortScience 40:529–531
Michelmore RW (2010) Genetic variation in lettuce. California leafy greens research program. http://calgreens.org/control/uploads/Michelmore_Variation_report_2009-2010_final_(2)1.pdf
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA (PNAS) 88:9828–9832
Michelmore RW, Truco MJ, Ochoa OE (2011) Breeding crisphead and leafy lettuce. California leafy greens research program. http://calgreens.org/control/uploads/Breeding_Crisphead_and_Leafy_Lettuce.pdf
Millani MJ, Erebarian HR, Alizadeh A (1999) Occurrence of fusarium wilt of lettuce in Shahr-Ray, Varamim and Karaj areas. Iran J Plant Pathol 35:44–45
Nagata RT (1992) Clip-and-wash method of emasculation for lettuce. HortScience 27:907–908
Paran I, Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985–993
Pasquali M, Dematheis F, Gilardi G, Gullino ML, Garibaldi A (2005) Vegetative compatibility groups of Fusarium oxysporum f. sp. lactucae from lettuce. Plant Dis 89:237–240
Pasquali M, Dematheis F, Gullino ML, Garibaldi A (2007) Identification of race 1 of Fusarium oxysporum f. sp. lactucae on lettuce by inter-retrotransposon sequence-characterized amplified region technique. Phytopathology 97:987–996
Ramalho MAP, Santos JB, Pinto CAB (2008) Genética na Agropecuária. 4th ed. Universidade Federal de Lavras, Lavras Minas Gerais State, MG, Brazil
Sala FC, Costa CD (2012) Retrospectiva e tendência da alfacicultura brasileira. Horticultura brasileira 30:187–194
Sala FC, Nascimento WM (2014) Produção de sementes de alface. In: Nascimento WM (ed). Produção de Sementes de Hortaliças. Embrapa Vegetable Crops, Brasília–DF, Brazil
Sandoya GV, Gurung S, Short DP, Subbarao KV, Michelmore RW, Hayes RJ (2017) Genetics of resistance in lettuce to races 1 and 2 of Verticillium dahliae from different host species. Euphytica 213:20
Santos JRM (1996) Methodology for screening tomato to Fusarium wilt, Verticillium wilt, gray leaf spot, early blight, and Septoria leaf spot. In: Proceedings of the international symposium on tropical tomato diseases. Recife, Pernambuco-PE, Brazil. pp 164–166
Schoeneweiss DF (1975) Predisposition, stress, and plant disease. Annu Rev Phytopathol 13:193–211
Schuster I, Cruz CD (2004) Estatística Genômica Aplicada a Populações Derivadas de Cruzamentos Controlados. UFV. Viçosa, Minas Gerais State, MG, Brazil
Scott JC, Kirkpatrick SC, Gordon TR (2010) Variation in susceptibility of lettuce cultivars to Fusarium wilt caused by Fusarium oxysporum f. sp. lactucae. Plant Pathol 59:139–146
Silva MP, Amaral AT Jr, Pereira MG, Rodrigues R, Daher RF, Posse SCP (2005) Diversidade genética e identificação de híbridos por marcadores RAPD em feijão-de-vagem. Acta Sci Agron 27:531–535
Simko I (2009) Development of EST-SSR markers for the study of population structure in lettuce (Lactuca sativa L.). J Hered 100:256–262
Simko I, Hayes RJ, Truco MJ, Michelmore RW (2011) Mapping a dominant negative mutation for triforine sensitivity in lettuce and its use as a selectable marker for detecting hybrids. Euphytica 182:157–166
Subbarao KV (1998) Progress toward integrated management of lettuce crop. Plant Dis 82:1068–1078
Tsuchiya N, Yoshida K, Usui T, Tsukada M (2004) Resistance tests and genetic resources for breeding Fusarium root rot resistant lettuce. J Jpn Soc Hortic Sci 73:105–113
Uwimana B, D’Andrea L, Felber F, Hooftman DAP, Den Nijs HCM, Smulders MJM, Visser RGF, Van De Wiel CCM (2012a) A Bayesian analysis of gene flow from crops to their wild relatives: cultivated (Lactuca sativa L.) and prickly lettuce (L. serriola L.), and the recent expansion of L. serriola in Europe. Mol Ecol 21:2640–2654
Uwimana B, Smulders MJM, Hooftman DAP, Hartman Y, Van Tienderen PH, Jansen J, Mchale LK, Michelmore RW, Visser RGF, Van De Wiel CCM (2012b) Crop to wild introgression in lettuce: following the fate of crop genome segments in backcross populations. BMC Plant Biol 12:43
Ventura JA, Costa H (2008) Fusarium wilt caused by Fusarium oxysporum on lettuce in Espírito Santo, Brazil. Plant Dis 92:976
Wang Y, Bao Z, Zhu Y, Hua J (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22:498–506
Whitham S, Mccormick S, Baker B (1996) The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc Natl Acad Sci USA 93:8776–8781
Yamauchi N, Shimazu J, Horiuchi S, Satou M, Shirakawa T (2004) Physiological races and vegetative compatibility groups of butterhead lettuce isolates of Fusarium oxysporum f. sp. lactucae in Japan. J Gen Plant Pathol 70:308–313
Zhu Y, Qian W, Hua J (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6:e1000844
Acknowledgements
Maria Esther de N. Fonseca, Ailton Reis, and Leonardo S. Boiteux were supported by fellowships from the Brazilian National Research Council (CNPq). Cléia Santos Cabral was supported by fellowships from CAPES and CNPq. The authors are also grateful to Dr. Elizabeth Georgian, Dr. Maria Josė Truco, and Dr. Richard W. Michelmore (UC Davis) for their careful review of the present manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have any conflicts of interest.
Informed consent
All authors have reviewed the manuscript and approved its submission to Euphytica.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cabral, C.S., Fonseca, M.E.N., Oliveira, V.R. et al. A single dominant gene/locus model for control of Fusarium oxysporum f. sp. lactucae race 1 resistance in lettuce (Lactuca sativa). Euphytica 215, 114 (2019). https://doi.org/10.1007/s10681-019-2441-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2441-2