Abstract
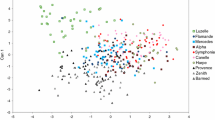
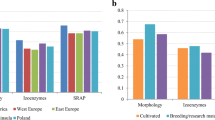
Feral populations of cultivated crops may act as reservoirs for novel traits and aid in trait movement across the landscape. Knowledge on the genetic diversity of feral populations may provide new insights into their origin and evolution and may help in the design of efficient novel trait confinement protocols. In this study, the genetic diversity of 12 feral alfalfa (Medicago sativa) populations originating from southern Manitoba (Canada) and 10 alfalfa cultivars and a M. falcata germplasm were investigated using eight SSR markers (i.e., microsatellites) and 14 phenotypic traits. We found that the genetic diversity observed in feral populations was similar to the diversity detected among the 10 cultivars. Analysis of molecular variance revealed that there was great genetic variation within (99.8%) rather than between different feral populations. Cluster analysis (unweighted pair-group method using arithmetic average) revealed no differentiation between feral populations and cultivars for neutral loci. High levels of population differentiation for phenotypic traits (and not for neutral markers) suggest the occurrence of heterogeneous selection for adaptive traits. The phenotypic traits we studied did not distinctly separate feral populations from cultivars but there was evidence of natural selection in feral populations for traits including winter survivability, rhizome production, and prostrate growth habit. Our results suggest that feral alfalfa populations need to be considered in the risk assessment of alfalfa containing novel genetically modified (GM) traits. Further, feral alfalfa populations may be regarded as a source of new germplasm for plant improvement.




Similar content being viewed by others
References
Bagavathiannan MV, Van Acker RC (2008) Crop ferality: implications for novel trait confinement. Agric Ecosyst Environ 127:1–6
Barnes DK, Bingham ET, Murphy RP, Hunt OJ, Beard DF, Skrdla WH, Teuber LR (1977) Alfalfa germplasm in the United States: genetic vulnerability, use, improvement, and maintenance. USDA-ARS Tech. Bull., Washington, DC, p 1571
Barton NH, Hewitt GM (1985) Analysis of hybrid zones. Annu Rev Ecol Syst 16:113–148
Berdahl JD, Wilton AC, Lorenz RJ, Frank AB (1986) Alfalfa survival and vigor in rangeland grazed by sheep. J Range Manage 39:59–62
Berdahl JD, Wilton AC, Frank AB (1989) Survival and agronomic performance of 25 alfalfa cultivars and strains inter-seeded into rangeland. J Range Manage 42:312–316
Bousquet J, Strauss SH, Doerksen AH, Price RA (1992) Extensive variation in evolutionary rate of rbcL gene sequences among seed plants. Proc Natl Acad Sci USA 89:7844–7848
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygous deficiency. Mol Ecol 5:453–455
Bruschi P, Vendramin GG, Bussotti F, Grossoni P (2003) Morphological and molecular diversity among Italian populations of Quercus petraea (Fagaceae). Ann Bot 91:707–716
Charlesworth D, Wright SI (2001) Breeding systems and genome evolution. Curr Opin Genet Evol 11:685–690
Cheung W, Hubert N, Landry B (1993) A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. PCR Methods Appl 3:69–70
Chiari Y, Hyseni C, Fritts TH, Glaberman S, Marquez C, Gibbs JP, Claude J, Caccone A (2009) Morphometrics parallel genetics in a newly discovered and endangered taxon of Galápagos tortoise. PLoS One 4:e6272
Conner JK, Hartl DL (2004) Primer to ecological genetics. Sinauer, Sunderland, Massachusetts, USA
Crochemore ML, Huyghe C, Ecalle C, Julier B (1998) Structuration of alfalfa genetic diversity using agronomic and morphological characteristics. Relationship with RAPD markers. Agronomie 18:79–94
Dakin EE, Avise JC (2004) Microsatellite null alleles in parentage analysis. Heredity 93:504–509
Diwan N, Bhagwat AA, Bauchan GR, Cregan PB (1997) Simple sequence repeat DNA markers in alfalfa and perennial and annual Medicago species. Genome 40:887–895
Excoffier L, Smouse PE, Quattro M (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Flajoulot S, Ronfort J, Baudouin P, Barre P, Huguet T, Huyghe C, Julier B (2005) Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a breeding program, using SSR markers. Theor Appl Genet 111:1420–1429
Fritz U, Siroký P, Hajigholi K, Michael W (2005) Environmentally caused dwarfism or a valid species—is Testudo weissingeri Bour, 1996 a distinct evolutionary lineage? New evidence from mitochondrial and nuclear genomic markers. Mol Phylogenet Evol 37:389–401
Gressel J (2005) Crop ferality and volunteerism. Taylor and Francis Publishing Group, Boca Raton, Florida, USA, p 422
Gutierrez-Ozuna R, Eguiarte LE, Molina-Freaner F (2009) Genetic diversity among pasture and roadside populations of the invasive buffelgrass (Pennisetum ciliare Link) in north-western Mexico. J Arid Environ 73:26–32
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Heinrichs DH (1963) Creeping alfalfas. Adv Agron 15:317–337
Husband BC, Barrett SCH (1991) Colonization history and population genetic structure of Eichhornia paniculata in Jamaica. Heredity 66:287–296
Jenczewski E, Angevain M, Charrier A, Ge’nier G, Ronfort J, Prosperi JM (1998) Contrasting patterns of genetic diversity in neutral markers and agro-morphological traits in wild and cultivated populations of Medicago sativa L. from Spain. Genet Sel Evol 30:5103–5120
Jenczewski E, Prosperi JM, Ronfort J (1999a) Evidence for gene flow between wild and cultivated Medicago sativa (Leguminosae) based on allozyme markers and quantitative traits. Am J Bot 86:677–687
Jenczewski E, Prosperi JM, Ronfort J (1999b) Differentiation between natural and cultivated populations of Medicago sativa (Leguminosae) from Spain: analysis with random amplified polymorphic DNA (RAPD) markers and comparison to allozymes. Mol Ecol 8:1317–1330
Julier B, Porcheron A, Ecalle C, Guy P (1995) Genetic variability for morphology, growth and forage yield among perennial diploid and tetraploid lucerne populations (Medicago sativa L). Agronomie 15:295–304
Julier B, Huyghe C, Ecalle C (2000) Within- and among-cultivar genetic variation in alfalfa: forage quality, morphology and yield. Crop Sci 40:365–369
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 3:9
Katepa-Mupondwa F, Singh A, Smith SR, McCaughey WP (2002) Grazing tolerance of alfalfa (Medicago spp.) under continuous and rotational stocking systems in pure stands and in mixture with meadow bromegrass (Bromus riparius Rehm. syn. B. biebersteinii Roem & Schult). Can J Plant Sci 82:337–347
Kendrick D, Pester T, Horak M, Rogan G, Nickson T (2005) Biogeographic survey of feral alfalfa populations in the U.S. during 2001 and 2002 as a component of ecological risk assessment of Roundup Ready® Alfalfa. In: Proceedings of north central weed science society meeting, UT, USA
Kruskal WH, Wallis WA (1952) Use of ranks on one-criterion variance analysis. J Am Stat Assoc 47:583–621
Lamari L (2002) Assess: image analysis software for plant disease quantification. The American Phytopathological Society Press, St Paul, MN, USA (118p + software CD)
Littel RC, Milliken GA, Stroup WW, Wofinger RD (1996) SAS system for mixed models. SAS Institute, Inc., Cary, NC
Lund RE (1975) Tables for an approximate test for outliers in linear models. Technometrics 17:473–476
Mahuku GS (2004) A simple extraction method suitable for PCR-based analysis of plant, fungal, and bacterial DNA. Plant Mol Biol Rep 22:71–81
Manske LL (2005) Evaluation of alfalfa varieties solid seeded into cropland. In: 2005 annual report, Dickinson Research Extension Center, ND. http://www.ag.ndsu.nodak.edu/dickinso/research/2004/range04i.htm . Accessed 08 Aug 2009
Melton B, Moutray JB, Bouton JH (1988) Geographic adaptation and cultivar selection. In: Hanson AA, Barnes DK, Hill RR Jr (eds) Alfalfa and alfalfa improvement. American Society of Agronomy, Inc. Publishers, Madison, Wisconsin, USA, pp 595–620
Pissard A, Arbizu C, Ghislain M, Faux AM, Paulet S, Bertin P (2008) Congruence between morphological and molecular markers inferred from the analysis of the intra-morphotype genetic diversity and the spatial structure of Oxalis tuberosa. Mol Genet 132:71–85
Prosperi JM, Jenczewski E, Angevain M, Ronfort J (2006) Morphologic and agronomic diversity of wild genetic resources of Medicago sativa L. collected in Spain. Genet Resour Crop Evol 53:843–856
Raybould AF, Mogg RJ, Aldam C, Gliddon CJ, Thorpe RS, Clarke RT (1998) The genetic structure of sea beet (Beta vulgaris ssp. maritime) populations. III. Detection of isolation by distance at microsatellite loci. Heredity 80:127–132
Rieseberg LH, Widmer A, Arntz AM, Burke JM (2002) Directional selection is the primary cause of phenotypic diversification. PNAS 99:12242–12245
Rohlf FJ (2000) NTSYS-PC numerical taxonomy and multivariate analysis system. Version 2.1. Exeter Publishing Ltd, Setauket, New York, USA
Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O’Neil P, Parker IM, Thompson JN, Weller SG (2001) The population biology of invasive species. Annu Rev Ecol Evol Syst 32:305–332
SAS Institute (2003) SAS 9.1 for windows. SAS Institute Inc, Cary, North Carolina, USA
Shaffer JP (1995) Multiple hypothesis testing. Annu Rev Psychol 46:561–584
Shannon LJ, Coll M, Neira S (2009) Exploring the dynamics of ecological indicators using food web models fitted to time series of abundance and catch data. Ecol Indic 9:1078–1095
Sledge MK, Ray IM, Jiang G (2005) An expressed sequence tag SSR map of tetraploid alfalfa (Medicago sativa L.). Theor Appl Genet 111:980–982
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463–6471
Thrall PH, Young A (2000) AUTOTET: a program for analysis of autotetraploids genotype data. J Hered 91:348–349
Van Acker RC (2007) The potential for the coexistence of GM and non-GM crops in Canada. In: Gulden RH, Swanton CJ (eds) The first decade of herbicide-resistant crops in Canada. Topics in Canadian Weed Science, vol 4. Canadian Weed Science Society, Sainte-Anne-de Bellevue, Québec, pp 153–162
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgments
This study was supported by Agri-Food Research and Development Initiative (ARDI) of the governments of Manitoba and Canada, the Natural Sciences and Engineering Research Council (NSERC) and a University of Manitoba Graduate Fellowship and Manitoba Graduate Scholarship for M. Bagavathiannan. Great thanks to INRA, Lusignan, France and also to the lab of Dr. Fouad Daayf, University of Manitoba, Canada, for providing the facilities to conduct the molecular work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bagavathiannan, M.V., Julier, B., Barre, P. et al. Genetic diversity of feral alfalfa (Medicago sativa L.) populations occurring in Manitoba, Canada and comparison with alfalfa cultivars: an analysis using SSR markers and phenotypic traits. Euphytica 173, 419–432 (2010). https://doi.org/10.1007/s10681-010-0156-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0156-5