Abstract
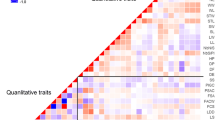
Diversity and structure of populations may differ substantially between morphological traits and molecular markers. Explanations of such discrepancies are crucial for further progress in breeding as well as for the maintenance of genetic resources. Our objective was to compare indices of among-cultivars differentiation for morphological traits (QST) and molecular markers (FST) in alfalfa (Medicago sativa), a legume forage species. Ten cultivars representing the Northern and Southern types commonly grown in Europe were investigated. For each cultivar, 40 genotypes were analysed with 16 SSR markers and four morphological traits measured in two locations and several cuts. QST values were in general high (0.02–0.39) compared to the differentiation observed with molecular markers (FST = 0.01), especially for growth habit, indicating that morphological traits were more efficient to structure the diversity than molecular markers. For morphological traits, a clear separation of Northern and Southern cultivars was observed, whereas for molecular markers, no clear structure was detected. Nevertheless, the grazing type cultivar Luzelle was separated from the rest of the cultivars for both morphological traits and molecular markers. Although pairwise differences between cultivars were significant for both morphological traits and molecular markers, the main part of the variation was found within cultivars. This large within-cultivar variation may be explained, besides the outcrossing reproductive mode and autotetraploid genetics, by the recent history of M. sativa domestication in Europe and the frequent seed exchanges. Selection for morphological traits (QST > FST) was achieved without modification of within-cultivar neutral diversity.

Similar content being viewed by others
References
Anderson JT, Willis JH, Mitchell-Olds T (2011) Evolutionary genetics of plant adaptation. Trends Genet 27:258–266. doi:10.1016/j.tig.2011.04.001
Annicchiarico P, Barrett B, Brummer EC, Julier B, Marshall AH (2015) Achievements and challenges in improving temperate perennial forage legumes. Crit Rev Plant Sci 34:327–380. doi:10.1080/07352689.2014.898462
Annicchiarico P, Nazzicari N, Ananta A, Carelli M, Brummer EC (2016) Assessment of cultivar distinctness in alfalfa: a comparison of genotyping-by-sequencing, simple-sequence repeat marker, and morphophysiological observations. Plant Genome 9:1–12. doi:10.3835/plantgenome2015.10.0105
Becker H (1993) Plant breeding. UTB Verlag Eugen Ulmer, Stuttgart
Bolaños-Aguilar ED, Huyghe C, Julier B, Ecalle C (2000) Genetic variation for seed yield and its components in alfalfa (Medicago sativa L.) populations. Agronomie 20:333–346
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygous deficiency. Mol Ecol 5:453–455
Cheung W, Hubert N, Landry B (1993) A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. PCR Methods Appl 3:69–70
Crochemore ML, Huyghe C, Kerlan MC, Durand F, Julier B (1996) Partitioning and distribution of RAPD variation in a set of populations of the Medicago sativa complex. Agronomie 16:421–432
Crochemore ML, Huyghe C, Ecalle C, Julier B (1998) Structuration of alfalfa genetic diversity using agronomic and morphological characteristics. Relationship with RAPD markers. Agronomie 18:79–94
Edelaar P, Burraco P, Gomez-Mestre I (2011) Comparisons between Q(ST) and F-ST-how wrong have we been? Mol Ecol 20:4830–4839
Endler JA (1977) Geographic variation, speciation, and clines vol 10. Monographs in population biology. Princeton University Press, Princeton
Flajoulot S, Ronfort J, Baudouin P, Barre P, Huguet T, Huyghe C, Julier B (2005) Genetic diversity among alfalfa (Medicago sativa) cultivars coming from a single breeding program, using SSR markers. Theor Appl Genet 111:1420–1429
Friedman J, Rubin MJ (2015) All in good time: understanding annual and perennial strategies in plants. Am J Bot 102:497–499. doi:10.3732/ajb.1500062
Herrmann D, Barre P, Santoni S, Julier B (2010) Association of a CONSTANS-LIKE gene to flowering and height in autotetraploid alfalfa. Theor Appl Genet 121:865–876. doi:10.1007/s00122-010-1356-z
Jenczewski E, Prosperi JM, Ronfort J (1999a) Differentiation between natural and cultivated populations of Medicago sativa (Leguminosae) from Spain: analysis with random amplified polymorphic DNA (RAPD) markers and comparison to allozymes. Mol Ecol 8:1317–1330
Jenczewski E, Prosperi JM, Ronfort J (1999b) Evidence for gene flow between wild and cultivated Medicago sativa (Leguminosae) based on allozyme markers and quantitative traits. Am J Bot 86:677–687
Julier B (1996) Traditionnal seed maintenance and origins of the French lucerne landraces. Euphytica 92:353–357. doi:10.1007/BF00037119
Julier B (2012) Alfalfa breeding benefits from genetic analyses on M. truncatula. In: Bushman S (ed) Molecular breeding of forage and turf, June 4–7, 2012. Utah State University, Utah, pp 17–19
Julier B, Porcheron A, Ecalle C, Guy P (1995) Genetic variability for morphology, growth and forage yield among perennial diploid and tetraploid lucerne populations (Medicago sativa L.). Agronomie 15:295–304
Julier B, Huyghe C, Ecalle C (2000) Within- and among-cultivar genetic variation in alfalfa: forage quality, morphology, and yield. Crop Sci 40:365–369. doi:10.2135/cropsci2000.402365x
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 3:9. doi:10.1186/1471-2229-3-9
Kidwell KK, Austin DF, Osborn TC (1994a) RFLP evaluation of nine Medicago accessions representing the original germplasm sources for North American alfalfa cultivars. Crop Sci 34:230–236
Kidwell KK, Bingham ET, Woodfield DR, Osborn TC (1994b) Relationship among genetic distance, forage yield and heterozygosity in isogenic diploid and tetraploid alfalfa populations. Theor Appl Genet 89:323–328
Kimura M (1983) Rare variant alleles in the light of the neutral theory. Mol Biol Evol 1:84–93
Le Corre V, Kremer A (2012) The genetic differentiation at quantitative trait loci under local adaptation. Mol Ecol 21:1548–1566
Leinonen T, O’hara RB, Cano JM, Merila J (2008) Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J Evol Biol 21:1–17
Li XH et al (2014a) Development of an alfalfa SNP array and its use to evaluate patterns of population structure and linkage disequilibrium. PLoS ONE. doi:10.1371/journal.pone.0084329
Li XH, Wei YL, Acharya A, Jiang QZ, Kang JM, Brummer EC (2014b) A saturated genetic linkage map of autotetraploid alfalfa (Medicago sativa L.) developed using genotyping-by-sequencing is highly syntenous with the Medicago truncatula genome. G3-Genes Genomes Genetique 4:1971–1979. doi:10.1534/g3.114.012245
Lonnet P (1996) Objectifs et critères actuels de la sélection des luzernes pérennes. Fourrages 147:303–308
Lynch M, Milligan BG (1994) Analysis of popualtion genetic structure with RAPD markers. Mol Ecol 3:91–99. doi:10.1111/j.1365-294X.1994.tb00109.x
Manninen O, Nissila E (1997) Genetic diversity among Finnish six-rowed barley cultivars based on pedigree information and DNA markers. Hereditas 126:87–93. doi:10.1111/j.1601-5223.1997.00087.x
Maureira I, Ortega F, Campos H, Osborn T (2004) Population structure and combining ability of diverse Medicago sativa germplasms. Theor Appl Genet 109:775–782
Mengoni A, Gori A, Bazzicalupo M (2001) Use of RAPD and microsatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Medicago sativa. Plant Breed 119:311–317
Muller MH, Prosperi JM, Santoni S, Ronfort J (2003) Inferences from mitochondrial DNA patterns on the domestication history of alfalfa (Medicago sativa). Mol Ecol 12:2187–2199
Musial JM, Basford KE, Irwin JAG (2002) Analysis of genetic diversity within Australian lucerne cultivars and implications for future genetic improvement. Aust J Agric Res 53:629–636
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prosperi JM, Jenczewski E, Angevain M, Ronfort J (2006) Morphologic and agronomic diversity of wild genetic resources of Medicago sativa L. collected in Spain. Genet Resour Crop Evol 53:843–856
Qiang H, Chen Z, Zhang Z, Wang X, Gao H, Wang Z (2015) Molecular diversity and population structure of a worldwide collection of cultivated tetraploid alfalfa (Medicago sativa subsp. sativa L.) germplasm as revealed by microsatellite markers. PLoS ONE 10:e0124592. doi:10.1371/journal.pone.0124592
Riday H, Brummer EC, Campbell TA, Luth D, Cazcarro PM (2003) Comparison of genetic and morphological distance with heterosis between Medicago sativa subsp. sativa and subsp. falcata. Euphytica 131:37–45
Rohlf RF (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system vol Exeter software, 2.1st edn. Applied Biostatistics Inc., New York
Roldan-Ruiz I et al (2001) A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor Appl Genet 103:1138–1150
Ronfort J, Jenczewski E, Bataillon T, Rousset F (1998) Analysis of population structure in autotetraploid species. Genetics 150:921–930
Segovia-Lerma AM, Cantrell RG, Conway JM, Ray IM (2003) AFLP-based assessment of genetic diversity among nine alfalfa germplasms using bulk DNA templates. Genome 46:51–58
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787–792. doi:10.1126/science.3576198
Small E (2011) Alfalfa and relatives: evolution and classification of Medicago. NRC Research Press of Canada, Ottawa
Spitze K (1993) Population structure in Daphnia obtusa—quantitative genetic and allozymic variation. Genetics 135:367–374
Thrall PH, Young A (2000) AUTOTET: a program for analysis of autotetraploid genotypic data. J Hered 91:348–349
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgements
We thank the ACVF (Association des Créateurs de Variétés Français) section Alfalfa for contributing to genotyping and phenotyping. JF Bourcier, F Durand, C Ecalle, A Gilly and J Jousse who contributed to genotyping or phenotyping are greatly acknowledged. This work was supported by Contrat de Branche from the French Ministry of Agriculture (2005–2008). DH received a grant from the Plant Breeding department of INRA.
Author information
Authors and Affiliations
Contributions
BJ, JR, CH conceived and designed the experiments; SF, BJ, PB performed the experiments; DH, BJ, SF analysed the data; DH, BJ, PB, JR, CH wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Herrmann, D., Flajoulot, S., Barre, P. et al. Comparison of morphological traits and molecular markers to analyse diversity and structure of alfalfa (Medicago sativa L.) cultivars. Genet Resour Crop Evol 65, 527–540 (2018). https://doi.org/10.1007/s10722-017-0551-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-017-0551-z