Abstract
Pectobacterium carotovorum subsp. odoriferum has been generally considered to have a narrow host range and has been isolated most often from chicory. Research was conducted to identify 91 Pectobacterium spp. strains isolated from different vegetables in Europe, North and South America, Asia, and Africa, and to compare their ability to cause disease in chicory and potato. Among the 91 strains, 22 strains from Europe were identified as P. c. subsp. odoriferum. Based on phylogenetic analysis of 16S rDNA, recA, and rpoS gene sequences, strains isolated from stored vegetables clustered together with the type strain of P. c. subsp. odoriferum and clustered separately from the P. c. subsp. carotovorum isolates. Eleven strains previously identified as P. c. subsp. carotovorum were reclassified as P. c. subsp. odoriferum. All P. c. subsp. odoriferum isolates were able to cause soft rot symptoms on chicory and potato. Moreover, the symptoms on potatoes were more severe at temperatures from 15 to 37 °C with P. c. subsp. odoriferum isolates than with P. atrosepticum or P. c. subsp. carotovorum isolates. Tissue maceration by P. c. subsp. odoriferum isolates was highest at 28 °C, and at that temperature tissue maceration was two-times greater for P. c. subsp. odoriferum isolates than for P. c. subsp. carotovorum isolates. Symptoms on inoculated chicory leaves were more severe with P. c. subsp. odoriferum (regardless of origin) than with other subspecies or species. To our knowledge, this is the first report that P. c. subsp. odoriferum occurs on a wide range of vegetables and has the ability to cause soft rot during potato storage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacteria belonging to the genus Pectobacterium cause many diseases on a great variety of crops worldwide. Pectobacterium spp. have been isolated from monocotyledonous and dicotyledonous plants, soil, and water, and have also been found in association with a variety of invertebrates (Glasner et al. 2008). Some species, such as P. atrosepticum, P. wasabiae, P. carotovorum subsp. carotovorum, and P. carotovorum subsp. brasiliense, cause diseases on a variety of crops but P. cacticida and P. betavasculorum have been isolated only from cacti and sugar beet, respectively (Alcorn et al. 1991; Gardan et al. 2003). The recently described species P. aroidearum preferentially infects monocots (Nabhan et al. 2013). P. carotovorum subsp. odoriferum has mostly been isolated from chicory in France (Samson et al. 1980; Gallois et al. 1992) and Japan (Lan et al. 2013) but can cause disease on hyacinth, celery, leek, and sugar beet (Gallois et al. 1992; Gardan et al. 2003). In recent years, strains of this subspecies were also isolated from okra in Malaysia (Nazerian et al. 2011), artichokes in Italy (Gallelli et al. 2009), Chinese cabbage in Korea (Thapa et al. 2011), and potato in Korea (Thapa et al. 2011) and Algeria (Yahiaoui-Zaidi et al. 2010). The identification of P. c. subsp. odoriferum has been based on RFLP analysis of pelABCDE genes, which encode pectate lyases (Darrasse et al. 1994) and 16S rRNA gene sequences (Hauben et al. 1998; Gardan et al. 2003). The identification of P. c. subsp. odoriferum has also been based on the recA gene (Waleron et al. 2002a), the rpoS gene (Waleron et al. 2002b), amplified fragment length polymorphism (AFLP) (Nabhan et al. 2012a), and multilocus sequence analysis (MLSA) (Ma et al. 2007; Kim et al. 2009; Nabhan et al. 2012b).
The first objective of this study was to identify newly isolated Pectobacterium strains and to verify the identities of Pectobacterium strains described in our previous work (Waleron et al. 2002a) in order to determine the presence of P. c. subsp. odoriferum on different vegetables. The second objective was to compare the ability of P. c. subsp. odoriferum isolated from chicory and other vegetables to induce disease symptoms on chicory leaves and potato tubers.
Materials and methods
Bacterial strains
Strains used in this study were listed in Table 1. Based on the research presented in this paper, the strains included three Pectobacterium species (P. atrosepticum, P. betavasculorum, and P. wasabiae) and three subspecies (P. c. subsp. carotovorum, P. c. subsp. odoriferum, and P. c. subsp. brasiliense). Among the 22 P. c. subsp. odoriferum strains, 11 were isolated in France, eight in Poland, two in Serbia, and one in Switzerland. All Polish and Serbian strains had been isolated from stored vegetables rather than from plants growing in the field.
Bacterial strains were grown in tryptic soy broth (TSB Bio-Merieux) shake culture at 28 °C for 24 h. For long-term storage, strains were kept in 40 % glycerol (v/v) at −80 °C.
Biochemical and physiological tests
The identities of the 91 Pectobacterium strains were carried out with the following biochemical analyses, which are routinely used to differentiate Pectobacterium subspecies (Schaad et al. 2001): indol production; phosphatase and urease activity; reducing substances from sucrose; acid production from maltose, rhamnose, trehalose, and α-methyl-D-glucoside; lactose fermentation; growth at 37 °C; growth on NA containing 5 % NaCl; L-lysine and L-ornithine decarboxylase activity, and L-arginine hydrolase activity. As described later, the identities were also assessed by molecular analysis.
Potato tuber assay
The ability of 87 Pectobacterium strains to macerate potato tubers (see Table 1) was determined using a potato tuber assay. Surface-sterilized potato tubers were inoculated by inserting one pipette tip containing 25 μl of bacterial suspension (2 × 107 cfu ml−1) 10 mm into each tuber as described previously (Lojkowska and Kelman 1994). The tubers were then incubated at 28 °C and 95 % relative humidity for 72 h before the diameter of the rotting tissue in each tuber was measured. For 15 P. c. subsp. odoriferum strains isolated from chicory and other vegetables, additional tubers were inoculated and incubated at 10, 15, 18, and 37 °C as before. Each combination of strain and temperature was represented by three replicate tubers, and the experiment was performed three times. As a negative control, pipette tips inserted into tubers contained sterile water rather than a bacterial suspension.
Chicory assay
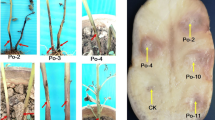
The ability to macerate chicory leaves was determined for 61 Pectobacterium strains (see Table 1). The inner surface of the chicory leaves was inoculated with 10 μl of a bacterial suspension (108 cfu ml−1), and the leaves were incubated in plastic bags at 28 °C and 95 % relative humidity for 24 h as described previously (Van Gijsegem et al. 2008). The 95 % humidity was created by placing a damp tissue in each bag. As a negative control, chicory leaves were inoculated with sterile water. Five leaves were inoculated per strain or control, and the experiment was performed twice. After 24 h at 28 °C, the length and width of the rotten tissue was measured on each leaf, and the average area of rotten tissue was calculated to estimate disease severity.
PCR amplifications, sequencing, and phylogenetic analysis
recA, rpoS, and 16S rRNA genes were amplified as described previously (Waleron et al. 2002a, 2008, 2013). The nucleotide sequences of recA, rpoS, and 16S rRNA genes were determined directly from PCR fragments amplified by the PCR primers used. The obtained sequences were deposited in GenBank under the following accession numbers: AY217078, AY217080, AY217082, AY217084, AY217086, AY264786, AY264789, AY264792, KC584977, KC584986, KC584989, KC584996, KC584997, and KF704760 to KF704816.
BLAST analysis was used to obtain sequences of recA, rpoS, and 16S rRNA genes from other Pectobacterium strains that were most similar to those of Pectobacterium carotovorum subsp. odoriferum. The sequences were obtained from the following nine Pectobacterium genomes: P. atrosepticum (BX950851, ASAB00000000), P. wasabiae (CP001790, CP003415, AKVS00000000), P. carotovorum (ABVY01000000, ABVX01000000, CP003776), and P. aroidearum (CP001657). The sequences were aligned using the MUSCLE algorithm with the default settings in Geneious Pro 6.1.6 (www.geneious.com).
The Maximum Likelihood (ML) phylogenetic analyses were performed with MEGA 5 software (www.megasoftware.net). Bootstrapping was executed with 1,000 replications. The recA, rpoS, and 16S rRNA gene sequences of Dickeya dadantii 3937 (CP002038) were used as outgroups.
Statistical analysis
The data were subjected to one-way ANOVAs using SPSS 10.0. Means were compared by the Tukey’s tests, and statistical significance was determined at 1 and 5 % levels.
Results
Phenotypic characterization and classification of bacterial isolates
All 91 strains were Gram-negative rods, facultatively anaerobic, and negative for oxidase and urease. All 91 strains degraded pectate, produced indol, liquefied gelatin, and grew at 28 °C. The strains isolated from different vegetables were further identified using PCR assays with primers specific for P. carotovorum (Darrase et al. 1994), P. atrosepticum (De Boer and Ward 1995), and Dickeya spp. (Nassar et al. 1996). All of tested strains except for P. betavasculorum generated PCR products with the P. carotovorum-specific primers (Darrase et al. 1994). None of the isolates was positive in the PCR assay with Dickeya spp.-specific primers (Nassar et al. 1996). Ten strains were identified as P. atrosepticum using PCR assay with primers specific for this species (De Boer and Ward 1995).
The biochemical properties of the eight strains, IFB5294-IFB5301, isolated from carrot, celery, leek, and onion in Poland, as well as two strains from parsley in Serbia (IFB5291, IFB5292) and one Swiss strain (IFB5293) from chicory, were identical with those observed for P. c. subsp. odoriferum strains isolated in France from chicory, celery, and leek (IFB5279-IFB5290), except for the lack of tolerance to 5 % NaCl in the medium.
All tested strains were negative for acid production from D-arabitol, dulicitol, and sorbitol. They were unable to utilize malonate, citrate, L-arginine, L-ornitine, and L-lysine. However, they were catalase positive and produced acid from lactose, maltose rhamnose, and trehalose. Also, they grew at 37 °C. The biochemical characteristics indicated that strains from Poland, Serbia, and Switzerland are P. c. subsp. odoriferum.
The abilities of P. c. subsp. odoriferum strains and strains from the other Pectobacterium species and subspecies to macerate potato tubers and chicory leaves were compared (Table 1). The mean diameter of rotting tissue in inoculated potato tubers was 18.4 mm for P. c. subsp. odoriferum, 17.2 mm for P. wasabiae isolated from potato, 7.8 mm for P. wasabiae isolated from horseradish and sweet pepper, 10.5 mm for P. betavasculorum, 11.6 mm for P. c. subsp. carotovorum, and 22.8 mm for P. c. subsp. brasiliense. Furthermore, the rotting in potato tubers caused by P. c. subsp. odoriferum was slightly higher for strains isolated from carrot, leek, parsley, and onion than for strains isolated from chicory (Table 2).
ANOVA and Tukey’s tests were used to compare the statistical significance of the observed differences in the ability to macerate of plant potato tubers tissues by strains belonging to different species and subspecies (Table 3). Maceration was significantly greater for strains belonging to the subspecies P. c. subsp. odoriferum than for P. betavasculorum strains and P. wasabiae strains isolated from horseradish. Moreover, maceration was significantly greater for P. c. subsp. odoriferum strains isolated from chicory than for P. wasabiae strains isolated from horseradish, and was greater for P. c. subsp. odoriferum strains isolated from vegetables other than chicory than for strains P. c. subsp. carotovorum strains, and P. wasabiae strains isolated from horseradish, but it was less than for P. c. subsp. brasiliense strains. Maceration was also greater for P. c. subsp. brasiliense stains than for strains of P. c. subsp. carotovorum, P. atrosepticum, and P. wasabiae (Table 3).
Maceration of potato tubers by 15 P. c. subsp. odoriferum strains was determined at 10, 15, 18, and 37 °C. None of 15 strains was able to macerate efficiently potato tubers at 10 °C (Table 4). All strains induced soft rot symptoms on potato tubers at temperatures from 15 to 37 °C. Tissue maceration was greatest at 28 °C. No maceration was observed on control water-inoculated control tubers.
All P. c. subsp. odoriferum strains in this study were able to macerate of chicory leave tissue. Maceration of chicory leaves was similar for P. c. subsp. odoriferum strains isolated from carrot, leek, parsley, and onion vs. P. c. subsp. odoriferum strains isolated from chicory (Table 1). Of the 61 Pectobacterium strains used in this experiment, the average value of the rotten area of chicory leaf tissue was highest for P. c. subsp. odoriferum strains (Table 1).
Molecular characterization of P. c. subsp. odoriferum strains
The 26 recA, 22 rpoS and 23 16S rDNA gene sequences were obtained. The sequences of P. c. subsp. odoriferum strains isolated from chicory were compared with those of P. c. subsp. odoriferum strains isolated from other Vegetables.
A Blast analysis of the 10 16S rRNA gene sequences obtained from P. c. subsp. odoriferum strains isolated from other vegetables (i.e., not from chicory) with those available in GenBank indicated that strains isolated from other vegetables exhibited 100 % identity with the type strain of P. c. subsp. odoriferum CFBP1878T and other P. c. subsp. odoriferum strains isolated from chicory.
The comparison of the recA and rpoS gene sequences of 14 of the 22 analyzed P. c. subsp. odoriferum strains showed that they were almost identical. Only one polymorphic position was observed in the amplified 735-bp fragment of recA, and two polymorphisms were observed in the 780-bp fragment of rpoS sequences.
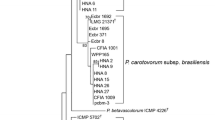
The topologies of maximum likelihood trees based on 16S rRNA (Fig. 1), recA (Fig. 2), and rpoS (Fig. 3) genes were congruent. The Swiss strain, as well as Polish and Serbian strains, isolated from different vegetables were clustered with the type strain of P. c. subsp. odoriferum CFBP1878T and other P. c. subsp. odoriferum strains isolated from chicory, and these clustered separately from the closely related subspecies P. c. subsp. carotovorum and P. c. subsp. brasiliense.
The genetic distance tree based on the 16S rRNA gene from Pectobacterium spp. Sequences obtained from the GenBank are described by accession numbers followed by strains numbers. Bootstrap values after 1,000 replicates are expressed as percentages. Bootstrap values < 50 % are excluded. Dickeya dadantii 3936 (CP002038) was included as an outgroup. The analysis used the consensus sequences of seven copies of the 16S rRNA gene from available genome sequences of: P. atrosepticum SCRI1043 (BX950851) and CFBP6276 (ASAB00000000); P. wasabiae CFBP3304 (AKVS00000000), WPP163 (CP001790), and SCC3193 (CP003415); P. c. subsp. carotovorum WPP14 (ABVY00000000) and PCC21 (CP003776); P. c. subsp. brasiliense (ABVX00000000); and P. aroidearum PC1 (CP001657). The consensus sequences are marked with stars
The consensus Maximum Likelihood tree based on the 735-bp fragment of the recA gene from Pectobacterium spp. Sequences obtained from the GenBank are described by accession numbers followed by strains numbers. Bootstrap values after 1,000 replicates are expressed as percentages. Bootstrap values < 50 % are excluded. Dickeya dadantii 3936 (CP002038) was included as an outgroup
The consensus Maximum Likelihood tree based on the 780-bp fragment of the rpoS gene from Pectobacterium spp. Sequences obtained from the GenBank are described by accession numbers followed by strains numbers. Bootstrap values after 1,000 replicates are expressed as percentages. Bootstrap values < 50 % are excluded. Dickeya dadantii 3936 (CP002038) was included as an outgroup
Discussion
In this study, we used several biochemical and molecular techniques to determine that P. c. subsp. odoriferum occurs on stored vegetables in Poland and Serbia. We have proved that the P. c. subsp. odoriferum subspecies was present in Poland already by 1995; however it was misidentified and classified as P. c. subsp. carotovorum. PCR-RFLP analysis of the recA gene was previously used to discriminate 18 different profiles among P. carotovorum strains (recA PCR-RFLP profiles 3–20) (Waleron et al. 2002a). Recently, strains assigned to recA PCR-RFLP profile 3 have been reclassified as P. wasabiae (Waleron et al. 2013). All P. c. subsp. odoriferum strains isolated from chicory were assigned to profile 22 recA PCR-RFLP (Waleron et al. 2002a). However, strains isolated from other vegetables and exhibiting biochemical characteristics typical of P. c. subsp. odoriferum strains were described as belonging to recA PCR-RFLP profile 13 (Waleron et al. 2002a). In the current study, the topologies of the maximum likelihood trees based on the 16S rRNA, recA, and rpoS genes clustered the Polish strains, one Swiss strain, and two Serbian strains, which belong to recA PCR-RFLP profile 13 (Waleron et al. 2002a) with P. c. subsp. odoriferum, and these strains clustered separately from the closely related subspecies P. c. subsp. carotovorum (Figs. 1, 2 and 3). The double-strand conformation polymorphism (DSCP) effect was the reason for assigning different recA PCR-RFLP profiles (13 and 22), for P. c. subsp. odoriferum strains (Waleron et al. 2002a) similarly as it was in case of recA PCR-RFLP profiles (3 and 23) for P. wasabiae strains from potato (Waleron et al. 2002a, 2013). According to our analysis of 16S rRNA, recA, and rpoS genes, those P. c. subsp. carotovorum strains that were previously identified as profile 13 recA PCR-RFLP (Waleron et al. 2002a) should be reclassified as P. c. subsp. odoriferum. Our results agree with MLSA studies of Nabhan et al. (2012b), which showed that Swiss strain 582 (IFB5293) is P. c. subsp. odoriferum. Although our study clearly indicates that P. c. subsp. odoriferum strains are genetically homogeneous, the P. c. subsp. odoriferum strains isolated from chicory were phenotypically slightly different from those isolated from other vegetables and caused less potato tuber maceration . However, it is too early to draw a general conclusion because only a limited number of strains were analyzed. The most significant difference was that strains from chicory but not from other vegetables could grow on a medium containing 5 % NaCl. Phenotypic diversity among P. c. subsp. odoriferum strains was also documented by Lan et al. (2013). The atypical phenotypic features of some P. c. subsp. odoriferum strains (e.g., resistance to 5 % NaCl in the medium) together with the DSCP effect observed in the RFLP analysis of the chosen genes could explain their misidentification in the past.
To our knowledge, this is the first report showing that P. c. subsp. odoriferum can cause soft rot of different vegetables during storage. Given that ‘Witloof’ chicory is grown underground or indoors in the absence of sunlight, the environment during chicory storage is similar to that during the storage of potatoes and other vegetables. We also note that the symptoms produced by P. c. subsp. odoriferum on stored chicory are similar to those produced on potato and other vegetables in storage.
Contrary to the results of Nabhan et al. (2012b), this study demonstrates that, at least under laboratory conditions, P. c. subsp. odoriferum strains are able to induce soft rot of potato tubers. In fact, the ability to macerate potato tuber tissues at 28 °C was greater for P. c. subsp. odoriferum (and also for P. c. subsp. brasiliense and P. wasabiae strains from potato) than for other Pectobacterium species and subspecies. However, 28 °C is higher than the optimal temperature for maceration by P. atrosepticum. Our results indicate that P. c. subsp. odoriferum subspecies are able to cause potato tuber tissue maceration like other potato-infecting species, e.g., P. atrosepticum, P. wasabiae, and subspecies P. c. subsp. carotovorum, which have been reported to cause disease on different plant species. Potato growers and processors should be concerned with the possibility that P. c. subsp. odoriferum could be transferred to potato tubers during storage. In the case of seed potatoes, the planting of the infected materials (obviously not tested for the presence of P. c. subsp. odoriferum) could result in losses. However it must be underlined that its ability to cause diseases symptoms on plants in field and storage conditions has to be confirmed.
P. c. subsp. odoriferum was first isolated in western Europe (Belgium, France, and Switzerland) (Gallois et al. 1992; Samson et al. 1980) and has since been reported in Poland, southern Europe (Gallelli et al. 2009), Africa (Yahiaoui-Zaidi et al. 2010), and Asia (Nazerian et al. 2011; Thapa et al. 2011; Lan et al. 2013). Although P. c. subsp. odoriferum strains have been isolated from potatoes in Korea (Thapa et al. 2011) and Algeria (Yahiaoui-Zaidi et al. 2010), the publications did not indicate when disease symptoms on potatoes were observed or the precise date of strain isolation.
P. c. subsp. odoriferum may also be present but incorrectly identified in other countries because distinguishing P. c. subsp. odoriferum from P. c. subsp. carotovorum is difficult. Methods relying on biochemical assays or restriction analysis of pel genes cannot always distinguish between these two groups. In addition, the use of MLSA does not always allow for the unambiguous identification of P. carotovorum (Nabhan et al. 2012b). The combination of biochemical and molecular methods allow for more effective differentiation of P. c. subsp. odoriferum from P. c. subsp. carotovorum.
In conclusion, the results presented here show that P. c. subsp. odoriferum strains can induce disease symptoms on potato and other vegetables in addition to chicory. Because P. c. subsp. odoriferum is widely distributed, its possible impact on vegetable production should be considered.
References
Alcorn, S. M., Orum, T. V., Steigerwalt, A. G., Foster, J. L. M., Fogleman, J. C., & Brenner, D. J. (1991). Taxonomy and pathogenicity of Erwinia cacticida sp. nov. International Journal of Systematic Bacteriology, 41, 197–212.
Darrasse, A., Priou, S., Kotoujansky, A., & Bertheau, Y. (1994). PCR and Restriction Fragment Length Polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Applied and Environmental Microbiology, 60, 1437–1443.
De Boer, S. H., & Ward, L. J. (1995). PCR detection of Erwinia carotovora subsp. atroseptica associated with potato tissue. Phytopathology, 85, 854–858.
Gallelli, A., Galli, M., De Simone, D., Zaccardelli, M., & Loreti, S. (2009). Phenotypic and genetic variability of Pectobacterium carotovorum isolated from artichoke in the Sele valley. Journal of Plant Pathology, 91, 757–761.
Gallois, A., Samson, R., Ageron, E., & Grimont, P. A. D. (1992). Erwinia carotovora subsp. odorifera subsp. nov., associated with odorous soft rot of chicory (Cichorium intybus L.). International Journal of Systematic Bacteriology, 42, 582–588.
Gardan, L., Gouy, C., Christen, R., & Samson, R. (2003). Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. International Journal of Systematic and Evolutionry Microbiology, 53, 381–391.
Glasner, J. D., Marquez-Villavicencio, M., Kim, H. S., Jahn, C. E., Ma, B., Biehl, B. S., et al. (2008). Niche-specificity and the variable fraction of the Pectobacterium pan-genome. Molecular Plant-Microbe Interactions, 21, 1549–1560.
Hauben, L., Moore, E. R. B., Vauterin, L., Steenackers, M., Mergaert, J., & Verdonck, L. (1998). Phylogenetic position of phytopathogens within the Enterobacteriaceae. Systematic and Applied Microbiology, 21, 384–397.
Kim, H. S., Ma, B., Perna, N. T., & Charkowski, A. O. (2009). Phylogeny and virulence of naturally occurring type III secretion system-deficient Pectobacterium strains. Applied and Environmental Microbiology, 13, 4539–4549.
Lan, W. W., Nishiwaki, Y., Akino, S., & Kondo, N. (2013). Soft rot of root chicory in Hokkaido and its causal bacteria. Journal of General Plant Pathology, 79, 182–193.
Lojkowska, E., & Kelman, A. (1994). Comparison of the effectiveness of different methods of screening for bacterial soft rot resistance of potato tubers. American Potato Journal, 71, 99–113.
Ma, B., Hibbing, M. E., Kim, H. S., Reedy, R. M., Yedidia, I., Breuer, J., Breuer, J., Glasner, J. D., Perna, N. T., Kelman, A., & Charkowski, A. O. (2007). The host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology, 97, 1150–1163.
Nabhan, S., De Boer, S. H., Maiss, E., & Wydra, K. (2012a). Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. Journal of Applied Microbiology, 113, 904–913.
Nabhan, S., Wydra, K., Linde, M., & Debener, T. (2012b). The use of two complementary DNA assays, AFLP and MLSA, for epidemic and phylogenetic studies of pectolytic enterobacterial strains with focus on the heterogeneous species Pectobacterium carotovorum. Plant Pathology, 61, 498–508.
Nabhan, S., De Boer, S. H., Maiss, E., & Wydra, K. (2013). Pectobacterium aroidearum sp. nov., a soft rot pathogen with preference for monocotyledonous plants. International Journal of Systematic and Evolutionary Microbiology, 63, 2520–2525.
Nassar, A., Darrasse, A., Lemattre, M., Kotoujansky, A., Dervin, C., Vedel, R., & Bertheau, Y. (1996). Characterization of Erwinia chrysanthemi by pectinolytic isozyme polymorphism and restriction fragment length polymorphism analysis of PCR- amplified fragments of pel genes. Applied and Environmental Microbiology, 62, 2228–2235.
Nazerian, E., Sijam, K., Ahmad, Z. A. M., & Keshavarz, K. (2011). Characterization of Pectobacterium carotovorum causing a new soft rot disease on okra in Malaysia. Journal of General Plant Pathology, 77, 292–294.
Samson, R., Poutier, F., Sailly, M., Hingand, L., & Jouan, B. (1980). Bactéries associées å une pourriture du collet de l’endive (in French). Annales de phytopathologie, 12, 311–317.
Schaad, N. W., Jones, J. B., & Chun, W. (2001). Laboratory guide for identification of plant pathogenic bacteria (3rd ed.). St. Paul: APS Press.
Thapa, S. P., Park, H. R., Lim, C. K., & Hur, J. H. (2011). Phylogeny of the Korean Erwinia species as determined by comparison of 16S rDNA sequences. Journal of Agricultural, Life and Environmental Sciences, 23, 62–69.
Van Gijsegem, F., Wlodarczyk, A., Cornu, A., Reverchon, S., & Hugouvieux-Cotte-Pattat, N. (2008). Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Molecular Plant-Microbe Interactions, 21, 1471–1481.
Waleron, M., Waleron, K., Podhajska, A. J., & Lojkowska, E. (2002a). Genotyping of bacteria belonging to the former Erwinia genus by PCR-RFLP analysis of recA gene fragment. Microbiology, 148, 583–595.
Waleron, M., Waleron, K., & Łojkowska, E. (2002b). Genotypic characterisation of the Erwinia genus by PCR-RFLP analysis of rpoS Gene. Plant Protection Science, 38, 288–290.
Waleron, M., Waleron, K., Geider, K., & Lojkowska, E. (2008). Differentiation of Erwinia amylovora and Erwinia pyrifoliae by PCR-RFLP of recA, gyrA and rpoS gene framents. European Journal of Plant Pathology, 121, 161–172.
Waleron, M., Waleron, K., & Lojkowska, E. (2013). Occurrence of Pectobacterium wasabiae in potato field samples. European Journal of Plant Pathology, 137, 149–158.
Yahiaoui-Zaidi, R., Ladjouzi, R., & Benallaoua, S. (2010). Pathogenic variability within biochemical groups of Pectobacterium carotovorum isolated in Algeria from seed potato tubers. International Journal for Biotechnology and Molecular Biology Research, 1, 1–9.
Acknowledgements
This work was supported by the National Science Centre (NCN N N310732240). The authors wish to thank two anonymous reviewers for their insightful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Waleron, M., Waleron, K. & Lojkowska, E. Characterization of Pectobacterium carotovorum subsp. odoriferum causing soft rot of stored vegetables. Eur J Plant Pathol 139, 457–469 (2014). https://doi.org/10.1007/s10658-014-0403-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-014-0403-z