Abstract
Anadromous salmonid fishes frequently exhibit strong geographic population structuring. However, population genetic differentiation of Atlantic salmon (Salmo salar) at fine geographic scales differs across equivalent spatial extents in different regions. So far, fine-scale genetic differentiation has not been assessed in rivers of the Baltic Sea, a region that contains an evolutionarily distinct Atlantic salmon lineage. Thus, Baltic salmon are currently managed on the river level, without focus on potential genetic structure and diversity within rivers. Here, we used microsatellites to characterize the genetic structure of wild juvenile salmon sampled throughout the interconnected, northern Baltic Tornio and Kalix Rivers. We found genetic differentiation within the two rivers, but not between them: salmon in the upper reaches differed from individuals in the lower reaches, regardless of river system. Further, examining smolts migrating from the river to the sea and adults returning from the sea to spawn, we found an association between the genetic structure and seasonal migration timing. Out-migrating smolts genetically assigned to upper river reaches were older and tended to reach the sea later in the season than smolts from the lower reaches. In contrast, mature adults originating from the upper reaches returned to the river early in the season. Our observation of genetic population structuring between downstream and upstream reaches of the large Tornio and Kalix rivers, and its association with migration timing, implies that careful temporal management of the northern Baltic fisheries would help to preserve the diversity and sustainability of the wild salmon stocks of these rivers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most species are spatially structured, with local populations linked by varying levels of connectivity. When gene flow between local populations is limited, genetic differences between populations arise and are maintained by random drift and local adaptation. Knowledge of genetic population structure is thus a general prerequisite for the identification of conservation and management units within species (e.g. Palsbøll et al. 2007; Funk et al. 2012; Östman et al. 2017).
Atlantic salmon (Salmo salar L. 1759, Salmonidae) is an anadromous fish, renowned for its long marine feeding migration and subsequent ‘homing’ back to its natal river to breed (Scheer 1939). This homing behaviour is reflected in genetically distinct populations among rivers (e.g. Ståhl 1987; O’Reilly et al. 1996; Fontaine et al. 1997; McConnell et al. 1997; Spidle et al. 2003; Verspoor et al. 2005; Griffiths et al. 2010). Even between neighbouring salmon rivers with only minor genetic divergence, levels of gene flow may be low enough to allow for a high degree of demographic independence (see Waples and Gaggiotti 2006). Rivers have therefore been considered as natural starting points when defining management units for Atlantic salmon (Dionne et al. 2009).
However, major river systems have been shown to host within-river genetic population structure. For example, Vähä et al. (2007, 2008) used microsatellites and found temporally stable genetic substructure in the large subarctic Teno River system, generating over 20 genetically distinct subpopulations of Atlantic salmon. These subpopulations were subsequently shown to harbour differentiation at the genomic level, associated with important life history diversity, including the age at which adults mature and return to their natal river (‘sea age’ or ‘age at maturity’), and the seasonal timing of when adult salmon enter the river to spawn (‘run timing’) (Pritchard et al. 2018). Substantial genetic population structure has also been found within the eastern Canadian Romaine, Moisie, Restigouche and Sainte-Marguerite Rivers (Garant et al. 2000; Landry and Bernatchez 2001; Dionne et al. 2009), the Moy and Foyle River systems in the island of Ireland (Dillane et al. 2007, 2008; Ensing et al. 2011), and the Penobscot River in Maine, USA (Spidle et al. 2001). Conversely, only weak genetic population structure occurs within the large Atlantic salmon stock spawning in the Miramichi River in eastern Canada (Dionne et al. 2009; Wellband et al. 2018). Furthermore, the Varzuga River in the Kola Peninsula in northern Russia shows weak within-river differentiation despite a clear pattern of ‘isolation-by-distance’ (IBD) (Primmer et al. 2006).
The long-term sustainability and ecosystem services of fish populations are buffered and enhanced by genetic and life history diversity, through a portfolio effect (Schindler et al. 2010). It is therefore important to maintain such diversity by avoiding the depletion of certain subcomponents of fish stocks (Hilborn et al. 2003; Hutchinson 2008; Vähä et al. 2017; Jacobson et al. 2019; Nordahl et al. 2019; Tamario et al. 2019). Thus, taking within-river population substructure into account can benefit salmon management, even though this can be difficult to achieve in practice (Potter et al. 2003). Population genetic studies are useful for this aim as they can help to identify the appropriate spatial scale at which to manage salmon, and thus to define conservation and management units and priorities (Dionne et al. 2009; Vähä et al. 2017).
The Baltic Sea supports a distinct evolutionary lineage of Atlantic salmon (Nilsson et al. 2001; Bourret et al. 2013). Wild Baltic salmon populations have been extirpated from almost three quarters of their native rivers due to the construction of hydropower dams, overfishing, habitat loss and pollution (e.g. Karlsson and Karlström 1994). Currently only 27 rivers out of around 90 in the Baltic contain self-sustaining salmon stocks considered to be wild (Koljonen 2001; ICES 2019). Earlier studies have found varying degrees of genetic differentiation among the remaining Baltic salmon rivers (e.g. Ståhl 1987; Koljonen et al. 1999; Säisä et al. 2005; Verspoor et al. 2005; Koljonen 2006), with a general pattern of IBD and three main population groups, assumed to mirror post-glacial colonization events from different refugia (Koljonen et al. 1999; Nilsson et al. 2001; Säisä et al. 2005; Tonteri et al. 2005).
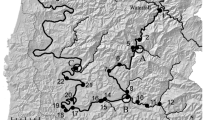
The neighbouring northern Baltic Rivers Tornio and Kalix are currently estimated to produce more than 70% of all wild Baltic salmon smolts (juveniles migrating from rivers to the sea) (ICES 2019). The Tornio River (Torne in Swedish; length 522 km, watershed area 40,157 km2, c. 50,000–150,000 returning spawners annually in recent years, ICES 2019) flows on the border of Finland and Sweden, with its mainstem and tributaries spanning both countries. The Kalix River (length 461 km, watershed area 23,600 km2, c. 30,000–60,000 returning spawners annually in recent years, ICES 2019) is located entirely in Sweden. These two unregulated major rivers have their mouths located just 50 km apart, at the northernmost rim of the Gulf of Bothnia. The rivers are connected by a large natural bifurcation (Tärendö River) where more than 50% of the annual discharge of the Swedish Torne River tributary flows into the Kalix River main stem (Fig. 1). As in most other Baltic rivers, the Tornio-Kalix salmon stock underwent a severe population decline over the twentieth century, and was considered close to extinction in the late 1980s (Pruuki 1993; Romakkaniemi et al. 2003). In response to this decline, the lower Tornio mainstem and the mid to upper Muonio tributary (Fig. 1) were heavily stocked between 1977 and 2002, with hatchery-reared 1st–3rd generation juvenile offspring of returning adults captured at the Tornio river mouth (Romakkaniemi et al. 2003; Anttila et al. 2008). Little or no supplementary stocking was performed in other parts of the Tornio or Kalix Rivers (Romakkaniemi 2008; ICES 2019). Since the late 1990s, the Tornio-Kalix salmon stock has recovered rapidly, but continues to be subject to harvest pressure from offshore, coastal and riverine fisheries (ICES 2019). Its importance as the main source of wild Baltic salmon means that sustainable management of the stock is essential. The cross-border nature of the river system, coupled with the bifurcation connecting the two rivers poses a special management challenge. However, the bifurcation has not, as of yet, been considered particularly important in the assessment (e.g. ICES 2019) or management of the stocks.
The Tornio and Kalix Rivers and their salmon stocks are of equivalent size and importance to the extensively studied and geographically close Teno River. However, so far no comprehensive genetic survey of juvenile salmon from different parts within both the Tornio and Kalix Rivers has been carried out, and currently salmon in the two rivers are assessed (e.g. ICES 2019) and managed separately with the assumption of no structuring within each river (Palm et al. 2020). Previous studies, focusing mostly on differences between the Tornio and Kalix and sampling only a few sites within the rivers, have reported minor genetic differences among salmon parr (juveniles feeding in freshwater), smolts, and adults sampled from the system (e.g. Ståhl 1981; Koljonen and McKinnell 1996; Koljonen et al. 1999; Nilsson et al. 2001; Säisä et al. 2005; Verspoor et al. 2005; Koljonen 2006, but see Jansson 1993).
Here, we apply 18 microsatellite markers to samples of salmon from different life stages, to investigate the fine-scale population genetic structure within the Tornio-Kalix River system. We further examine whether this genetic structure is associated with life history variation in freshwater and marine life history traits.
Materials and methods
Samples
Parr: A total of 772 salmon parr were sampled between August and October in 2012 (n = 725) and 2013 (n = 47) at 45 electrofishing locations in all major branches of the Tornio-Kalix River system, including the bifurcation (Tärendö River, Fig. 1). For all sites except T2, fin-clips of sampled parr were taken in the field and stored in 95% ethanol in individual tubes. For site T2, scale samples were taken in the field and stored in paper envelopes. To allow for genetic comparisons of different age groups, tissue samples from yearlings (age 0+ , total length 30–60 mm) and older parr (age > 0+, total length > 60 mm) were collected as evenly as possible. The proportions of yearlings and older parr in the total material were 38.1% and 61.9%, respectively. For statistical analyses, the 45 locations were combined into 16 broader sampling sites based on geographical proximity and genetic homogeneity among locations within sites (results not shown) (Fig. 1).
Smolts: Tissue samples were collected from out-migrating smolts in 2011, using a trap placed close to the Tornio River mouth (Fig. 1). The trap was operated during the whole smolt migration season as part of a long-term monitoring program (starting 1 week after local ice break-up and ending when the daily smolt captures fell to below 0.1% of the total captures). Smolts from all upstream production areas were therefore assumed to be included in the collection. Samples were collected from every 320th trapped individual to a maximum of 5 fish per day from May 14th to June 28th, making a total of 196 smolts (107 females, 89 males). All smolts genotyped in this study had been killed at capture, measured, weighed, sexed, and subsequently aged by trained experts on the basis of scale growth rings, following international guidelines for Atlantic salmon scale reading (ICES 2011).
Adults: We genetically analyzed scale samples from 287 ascending adults that were caught by anglers 110–180 km from the Tornio River mouth (Fig. 1). The samples were collected in 2009 (87 females, 52 males, 6 unreported sex) and 2010 (106 females, 34 males, 2 unreported), throughout the entire fishing season (June 4th to August 15th). A maximum of 21 individuals (mean 4) per day were collected. Adults had been killed at capture, measured, weighed and sexed, and life history data (age of smolting in years, age at maturity in sea winters, number of previous spawnings) had been determined from scale growth rings as above (ICES 2011).
Microsatellite analyses
Total DNA was extracted following a Chelex extraction protocol (Walsh et al. 1991). The following 18 microsatellite loci were genotyped in two multiplexes: Ssa407 (Cairney et al. 2000), SSsp3016 (Gilbey et al. 2004), SSaD157 (King et al. 2005), Ssa14, Ssa289 (McConnell et al. 1995), Ssa85, Ssa171, Ssa197, Ssa202 (O’Reilly et al. 1996), SSsp1605, SSsp2201, SSsp2210, SSsp2216, SSspG7 (Paterson et al. 2004), SsOsl85, SsOsl311, SsOsl417, and SsOsl438 (Slettan et al. 1995). For each PCR reaction we used 4 µL of Type-it Multiplex PCR Master Mix (Qiagen), 4 µL multiplex primer mix, and 0.6 µL template with approximately 100 ng DNA. Uniform signal intensity among loci was achieved by adjusting primer concentrations. PCR was run with an initial step of 5 min at 95 °C followed by 25 cycles of 30 s at 95 °C, 90 s at 56 °C, 30 s at 72 °C and a final step of 15 min at 60 °C. Electrophoresis was performed on an ABI 310 with Liz 500 size standard and allele sizes were determined using the ABI Genotyper 3.7 software.
Statistical analyses
In order to avoid potentially biased allele frequency estimates due to family sampling of juvenile salmon (Hansen et al. 1997; Östergren et al. 2020), Colony 2.0.6.5 (Wang 2004; Jones and Wang 2010) was used to identify full siblings within the parr dataset. A total of 16 analyses (corresponding to the 16 sample sites) were performed with Colony, assuming polygamous mating patterns in both sexes as is typical for salmonids (Fleming 1996). We used the following threshold parameters for identifying full-sibs: Best (ML) full-sib family Prob. (Inc.) > 0.90 AND Prob. (Exc.) > 0.90. We retained only one parr from each identified full-sib family for the analyses below.
The programs Genepop 4.7.3 (Rousset 2008), Arlequin 3.5.1.2 (Excoffier and Lischer 2010) and Fstat 2.9.4 (Goudet 2003) were used for computing population genetic parameters. Exact tests in Genepop were used to identify statistically significant deviations from Hardy–Weinberg proportions across loci (within sites) and sites (within loci). We also used Genepop to identify deviations from genotypic linkage equilibrium, in all pairs of loci and within all sites. We used the Markov chain parameters of 10,000 steps of dememorization, 1000 batches and 10,000 iterations per batch for all analyses, and assessed the significance of the results by applying the Bonferroni correction for multiple tests (Rice 1989).
Arlequin was used for computing the number of alleles per population and loci, F-statistics (global and pairwise FST estimates between sites, and significance assessed with 100,000 permutations; Weir and Cockerham 1984), and for performing an analysis of molecular variance (AMOVA) to assess genetic differences within versus between the rivers, with 100,000 permutations. Fstat was used to compute allelic richness (locus allele number standardized to the smallest sample size of 25 individuals).
Genetic isolation-by-distance (IBD) among parr samples was analyzed using the Mantel test with 100,000 permutations in Arlequin, with FST/(1 − FST) as genetic distance and the shortest waterway (either via the bifurcation or the sea) as geographical distance between the sites.
We used Poptreew (Takezaki et al. 2014) to estimate Nei’s DA (Nei et al. 1983) between the 16 parr sampling sites, for constructing a neighbour-joining tree (Saitou and Nei 1987), and estimated node support using 5000 bootstraps (Felsenstein 1985). We edited the tree with FigTree 1.4.3 (Rambaut 2012).
We used Structure 2.3.4 (Pritchard et al. 2000; Falush et al. 2003) to examine population genetic structure across the two rivers. All Structure models (burn-in of 50,000 steps, followed by 100,000 MCMC replicates) were run without prior information about sampling locations, and assuming admixture and correlated allele frequencies between genetic clusters. The number of clusters (K) was increased from 1 to 16 (for the total parr material, and separate runs for the two different parr age classes), or from 1 to 5 (for the total parr, smolt and adult material), always with 5 replicate runs per K. True K was inferred following Evanno et al. (2005), as implemented in Structure harvester (Earl and vonHoldt 2012), and the results were visualized by Clumpak 1.1 (Kopelman et al. 2015). Because studies relying on the ΔK method may show a dramatic overrepresentation of K = 2 (Janes et al. 2017; Cullingham et al. 2020), we also examined the maximum likelihood of K for each value of K (Ln Pr(X|K)) with Structure harvester.
We performed genetic mixed stock analyses (MSA) for the Tornio River smolts and adults using the R (R Core Team 2020) package rubias (Moran and Anderson 2019). Because of the bifurcation between the Tornio and Kalix Rivers, smolts and adults sampled in the lower Tornio could potentially originate from spawning areas in either the Kalix or the Tornio. We therefore used all 16 parr sampling sites as a genetic baseline, and split them to two reporting groups, ‘Lower’ and ‘Upper’ reaches (Table 1), based on geographical location and the results of our population genetic analyses (see below). First, we used a leave-one-out reassignment procedure (Anderson et al. 2008) to evaluate expected accuracy of assignment to the 16 parr sites and two reporting groups. Each fish was assigned to the site to which it had the highest posterior probability of assignment (‘scaled likelihood’ in rubias), and reporting group accuracy was estimated from combined site assignments. Subsequently, we performed separate genetic mixture analyses for the smolts and adults to estimate the proportion of the total mixture originating from each parr site and reporting group, and the posterior mean of reporting group membership for each individual. We qualitatively explored changes over the sampling season by dividing the data into 1 week intervals for smolts and 2 week intervals for adults and repeating the mixture analysis (see above) for these subsets. We used the default rubias maximum likelihood approach with 2000 MCMC sweeps and discarded 200 sweeps as burn-in.
We used Chi squared tests to test for independence between sex, sea age, and smolt age, using the categories described below. To investigate whether there were differences in smolt age or sea age between different parts of the river system, we first performed multinomial logistic regression using the nnet package in R (Venables and Ripley 2002), with each individual’s posterior mean of membership to the ‘Upper’ reporting group (hereafter ‘P(Upper)’) as the explanatory variable. For smolt age (assessed from both smolts and adults), we defined three categories for the response variable: 2 years (n = 31), 3 years (n = 289), and 4–5 years (n = 162). For sea age (assessed from adults only), we defined five categories for the response variable: 1 sea winter (1SW; 16 males, 1 female, 1 unknown), 2 sea winter (2SW; 45 males, 114 females, 5 unknown), 3 sea winter (3SW; 19 males, 31 females, 1 unknown), 4–5 sea winter (4–5SW; 15 females, 2 males) and repeat spawners (RS; fish that had returned to spawn at least once previously, independent of their age at first return; 4 males, 32 females, 1 unknown). We assessed statistical significance using a two-tailed z test.
Because of the highly skewed nature of our smolt age dataset (both in terms of age and P(Upper)), we further investigated the robustness of any relationship between smolt age and P(Upper) using a randomization approach. We created 5000 datasets with no association between P(Upper) and smolt age by assigning each individual a random age generated by sampling with replacement from the true age distribution. We calculated median P(Upper) for each of the three smolt age categories for each of the 5000 randomized datasets, and compared the true median P(Upper) to the distribution of the simulated median P(Upper).
To investigate the relationship of P(Upper), smolt age (smolts only), sea age (adults only), year (adults only), and sex (smolts and adults) to seasonal migration timing, we used the MASS package in R (Venables and Ripley 2002) to fit a negative binomial generalized linear model (GLM with a log link) to catch date (coded as number of days since May 1 for smolts, and days since June 1 for adults). In order to compare the Akaike information criterion (AIC) between models with the same number of measurements for each variable, we removed 8 individuals with unknown sex. We included explanatory variables sea age, sex and year as fixed effects without interactions in the model. We then simplified the model by stepwise reduction (using the step function in R), to select the model with the lowest AIC.
Again, because of the highly skewed nature of our smolt dataset in terms of P(Upper), we further investigated the robustness of any relationship between P(Upper) and smolt capture date using a randomization approach. Because running large numbers of GLMs would be inefficient, we instead used the Spearman rank correlation coefficient (ρ) between the two variables as our exploratory measure. To retain the temporal structure of our dataset (maximum 5 individuals sampled per day), we created 5000 datasets by assigning each individual a random P(Upper) generated by sampling with replacement from the true P(Upper) distribution. We compared the true ρ to the distribution of ρ across the simulated datasets.
Results
Genetic variation
Colony identified 34 putative full-sibs from 11 families (7 families with 2 full-sibs, 1 family of each of the following full-sib family sizes: 3, 4, 5 and 8). Removal of all but one individual in each full-sib family resulted in a final total parr sample of 749 individuals.
No parr sampling sites (i.e. combinations of nearby electrofishing locations) or microsatellite loci (across sites) deviated significantly from Hardy–Weinberg genotypic proportions after Bonferroni correction for multiple tests (α = 0.05) (Table 1; Online Resource 1, Table S1). Furthermore, none of the loci showed consistent signs of linkage after pairwise tests within sites and Bonferroni correction (α = 0.05) (not shown). Therefore, we retained all 18 microsatellite loci in our analyses.
Number of alleles per locus observed in all samples combined (parr, smolts, adults; n = 1232) ranged from 3 to 29 (mean 14.6). Mean HE across loci was 0.71, with a range from 0.21 to 0.93 (Online Resource 1, Table S1), and 0.70 over all sites, ranging from 0.67 in K4 (Kaitum River) to 0.73 in T7 (Muonio River) (Table 1). Mean HE in the ‘Upper’ reporting group of the Kalix River system ranged from 0.67 to 0.69, and from 0.70 to 0.72 in the ‘Lower’ reporting group. Mean HE in all sites of the ‘Upper’ reporting group of the Tornio River system was 0.70, and ranged from 0.70 to 0.73 in the ‘Lower’ reporting group. Mean allele richness was 8.11, ranging from 6.84 in K4 (Kaitum River) to 8.98 in T2 (Tornio River) (Table 1).
Within-river genetic structure
Global mean FST among sampling sites was low but statistically significant, both across and within rivers (Both rivers: FST=0.015, P < 0.001; Kalix only: FST = 0.015, P < 0.001; Tornio only: FST = 0.015, P < 0.001). A hierarchical AMOVA revealed no overall genetic differentiation between salmon parr in the Kalix and Tornio rivers (Fbetween rivers = − 0.0004, P = 0.411), but clear differentiation among sites within the two rivers (Fwithin rivers= 0.015, P < 0.001) (Online Resource 2, Table S2).
Pairwise FST values between the 16 parr sites are shown in Table 2. Out of 120 pairwise comparisons, 90 (75%) were statistically significant (P < 0.0004) following Bonferroni correction (α = 0.05). Low and non-significant pairwise FST values were observed among samples from the lowest parts of the two rivers (sites K1, K2, T1, T2 and T3; 9 of 10 comparisons non-significant). The highest estimated FST (0.062) was found between sites K4 and K6 from different branches of the Kalix River system (tributaries Kaitum River and Ängesån River, respectively). Parr from the tributaries Ängesån River (sites K6 and K7) and Lainio River (T6) differed significantly from parr from all other sites.
A neighbour-joining tree based on Nei’s genetic distance (Nei et al. 1983) further supported the occurrence of larger genetic differences within than between the rivers, with upstream and downstream samples tending to be more genetically similar to one another regardless of river (Fig. 2). Strongest bootstrap supports (77 to 93%) were found for branches with the sites K6 and K7 (Kalix River tributary Ängesån River), three upstream Kalix River sites K3-K5, and a larger group including the upstream Kalix samples (K3-K5) and all three uppermost sites from the different Tornio River tributaries (T4, T6 and T8, Fig. 2).
Unrooted neighbour-joining dendrogram based on Nei’s DA among the 16 sample sites in the Tornio-Kalix River system, with the ‘Lower’ (downstream) and ‘Upper’ (upstream) reporting groups illustrated. Bootstrap support values ≥ 75% are shown. Pie charts illustrate mean ancestry from the two genetic clusters identified using Structure
We observed a pattern of isolation-by-distance (IBD) when measuring distance among all sampling sites either via the bifurcation (r = 0.487, P <0.001, Fig. 3), or via the sea (r = 0.226, P = 0.044). Signals of IBD were also obtained when performing the analysis separately within each river (Kalix, bifurcation excluded: r = 0.721, P = 0.005; Tornio: r = 0.492, P =0.046).
Structure results suggested K = 2 genetic clusters, both within the sampled parr and for all sampled fish (parr, smolts and adults combined), supported by Ln Pr(X|K) values and/or the ΔK statistic (Fig. 4; Online Resource 3, Fig. S1, S2, Table S3). The clusters did not correspond to the two rivers; instead, parr from the lower parts of both rivers exhibited more ancestry from one cluster, and parr from the upper parts of both rivers more ancestry from the second cluster (Fig. 4). Separate Structure runs for parr of different ages produced similar results supporting K = 2 (Online Resource 3, Fig. S3).
Inferred ancestry of salmon parr from the Kalix (K1–K8) and Tornio Rivers (T1–T8) to two genetic clusters (K = 2) according to Structure. Each vertical line represents an individual fish. Horizontal lines underneath the codes for sampling sites illustrate ‘Lower’ (downstream) and ‘Upper’ (upstream) reporting groups
Mixture analysis
In the baseline reassignment test, 84.8% of parr from the ’Lower’ reaches and 78.8% from the ’Upper’ reaches of the Tornio-Kalix system were reassigned to their correct reporting groups. Estimated P(Upper) for each of the baseline individuals, calculated as the sum of posterior assignment probabilities to each of the ‘Upper’ collection sites, is shown in Fig. 5. Mixture analysis inferred the majority of sampled smolts and adults to originate from sites in the ‘Lower’ reporting group (estimated mixture proportions from ‘Upper’ reporting group: Smolts 2011, mean = 0.066, 95% CI 0.021–0.121; Adults 2009, mean = 0.294, 95% CI 0.213–0.385; Adults 2010, mean = 0.450, 95% CI 0.356–0.543).
Assignment accuracy at the level of the 16 parr sites was much lower than for the two reporting groups, with > 50% of baseline individuals correctly reassigned only for sites K4, K7, T6 and T8 (Online Resource 4, Table S4). Due to this, we collapsed the individual sites to the ‘Lower’ and ‘Upper’ reporting groups (Online Resource 4, Fig. S4, S5). In both years, sites from the ‘Upper’ reporting group contributed a large proportion of the earliest returning adults. The large majority of smolts appeared to originate from sites in the ‘Lower’ reporting group (Online Resource 4, Fig. S4).
Smolt age (examined in smolts and adults) was independent of sex, but sea age (examined in adults only) was not: 1SW fish were almost exclusively males, while 4–5SW fish and repeat spawners were disproportionately female (sex vs. smolt age: χ2 = 3.84, df = 2, P = 0.147; sex vs. sea age: χ2 = 43.078, df = 4, P < 0.0001). Multinomial logistic regression demonstrated P(Upper) to be related to smolt age: fish with a higher P(Upper) tended to spend a longer time in freshwater before smolting (Table 3, Fig. 6a; Online Resource 4, Fig. S6a). This relationship was further confirmed by the randomization analysis: observed median P(Upper) for 2 year and 3 year old smolts was in the lowest part of the simulated neutral distribution, while for 4–5 year old smolts it was higher than any simulated value (2Y, median P(Upper) = 0.0006, P = 0.0076; 3Y, median P(Upper) = 0.0083, P = 0.0426; 4–5Y, median P(Upper) = 0.1200, P < 0.0002). P(Upper) was also related to sea age, which was driven by almost all of the 1SW adults having a very low probability of assignment to the ‘Upper’ reporting group. P(Upper) was not related to age of return for fish older than 1SW (Table 3, Fig. 6b; Online Resource 4, Fig. S6b).
a Seasonal timing of out-migrating smolts from the Tornio River in 2011. Capture date shows days since May 1. P(Upper) shows each individual’s posterior mean of membership to the ‘Upper’ reporting group. Each panel includes original data (data points) and model predicted values with 95% confidence bands. Membership to the ‘Upper’ reporting group significantly delays capture date (see Table 4). b Seasonal timing of returning adults caught in the Tornio River in 2009 and 2010. Capture date shows days since June 1. P(Upper) shows each individual’s posterior mean of membership to the ‘Upper’ reporting group. The panels include original data (data points) and model predicted values for different age classes, with 95% confidence bands. Genetic cluster, sea age and year significantly affect capture date (see Table 4)
The negative binomial GLM that provided the best fit for smolt capture date included only P(Upper) as an explanatory variable. Smolts with a higher P(Upper) arrived at the river mouth later in the smolt migration season (Table 4, Fig. 6a). The randomization analysis examining Spearman rank correlation coefficient between P(Upper) and smolt capture date supported this relationship: observed ρ (0.358) was higher than all 5000 simulated ρ (P < 0.0002). The model that provided the best fit for adult catch date included the following explanatory variables: year, sea age, and P(Upper) (Table 4). In both years, adult salmon returning earlier in the season had a higher P(Upper) than individuals returning later. The median catch dates of 2–5SW fish (19 days since June 1) and repeat spawners (13 days since June 1) were also early compared to 1SW adults (median catch date 56.5 days since June 1), none of which were caught in the first half of the fishing season (Fig. 6b). However, this later migration date of 1SW fish was essentially based on data from 2009 only, as only one 1SW individual was sampled in 2010.
Discussion
We found population genetic structuring within the wild Atlantic salmon stocks of the Tornio and Kalix Rivers in the northern Baltic. Interestingly, we did not find evidence of overall genetic divergence between these two large interconnected rivers, but rather parallel genetic structuring within each one. In the Tornio River, this internal structure was associated with differences in migration timing of emigrating smolts and adults returning to the river to spawn. The link between genetic structure and observed life history variation in the river system indicates that our findings are important for the management of these largest remaining wild Baltic salmon rivers.
Upstream–downstream isolation-by-distance within rivers
We observed clear genetic differentiation between the upstream and downstream sections of both the Kalix and the Tornio Rivers, associated with an underlying pattern of isolation-by-distance (IBD). By comparing parr of different ages we found that this genetic structure was stable across at least two years. Similar upstream–downstream structuring in the Kalix was also observed in the 1980s by Jansson (1993). Additionally, the Jokkfall waterfall (just north of site K2) in the Kalix River has been a natural, partial migration obstacle that salmon could pass only during low water flow, until a fish ladder was built beside it in 1980 (Jansson 1993). This historical barrier has likely contributed to the upstream–downstream genetic differentiation in the Kalix. Furthermore, the presence of a strong IBD implies population genetic stability over a longer period of time, as has been found in other large Atlantic salmon populations (Vähä et al. 2008; Ozerov et al. 2013). It is unclear whether the identification of two genetic clusters by the Bayesian clustering algorithm indicates a true genetic transition zone within the rivers, or if it merely reflects an artefact caused by the IBD pattern (Frantz et al. 2009; Perez et al. 2018). However, both scenarios support a model of fine-scale natal homing with salmon returning to relatively specific spawning sites within the river system.
Large-scale supplementary stocking using salmon of local origin took place in the Tornio River system from 1977 to 2002 (close to sites T1, T2, T7 and T8; Romakkaniemi 2008). Although extensive, these releases have not been considered a key factor in the population recovery of the Tornio-Kalix salmon (Romakkaniemi et al. 2003). This past stocking has apparently not entirely obscured genetic differentiation in the system, despite its potential to artificially increase gene flow between the upper and lower reaches. However, it is still unknown if within-river genetic structuring has decreased over time. Genetic analysis of historic samples collected before the stocking activities would clarify the effects of this practice.
Our results underline the difficulty of generalizing genetic within-river structuring of large Atlantic salmon populations (see Dionne et al. 2009). The multiple large tributaries of the Tornio-Kalix River system (Fig. 1, global FST = 0.015) might be expected to host strongly differentiated subpopulations, as in the geographically adjacent Teno River (Vähä et al. 2007, 2008, global FST= 0.067). Instead, the amount of genetic differentiation among sampling sites and the IBD pattern observed in the Tornio and Kalix mirror that seen in the large Varzuga River system of northern Russia (Primmer et al. 2006, global FST = 0.014), and in the Teno River main stem (Vähä et al. 2017). Conversely, the degree of population genetic structuring in the Tornio and Kalix is higher than that of the large Miramichi system and salmon stock in Canada (global FST = 0.004; Dionne et al. 2009; Moore et al. 2014; Wellband et al. 2018).
The relatively low level of genetic substructure observed at the present microsatellite markers does not rule out the possibility of adaptive genetic differentiation in the Tornio-Kalix system at other loci. For example, by using a genomic dataset, Wellband et al. (2018) found support for adaptive processes associated with summer precipitation being the primary force in shaping the genetic structure in the Miramichi system, with very low population differentiation at presumably neutral markers (Dionne et al. 2009; Wellband et al. 2018). Despite weak neutral differentiation, the tributaries and different parts of the Tornio-Kalix system may contain adaptive variation associated with environmental characteristics. One possible selective agent is the Atlantic salmon flatworm parasite Gyrodactylus salaris. Lumme et al. (2016, see also Anttila et al. 2008; Kuusela et al. 2009) found stable upstream–downstream genetic divergence of G. salaris in the Tornio, and coadaptation between the local parasite populations and local salmon host populations is one mechanism that could maintain upstream–downstream structuring in the river by reducing the fitness of inter-population hybrids (see Karvonen and Seehausen 2012).
Genetic homogeneity between rivers
In contrast to the upstream–downstream structure in the Kalix and Tornio, we observe little overall genetic differentiation between the two rivers. Instead, downstream sites in one river are genetically more similar to downstream sites in the other river than to upstream sites in the same river, and vice versa. This suggests the occurrence of homogenizing gene flow between equivalent lower and upper sections of the two different rivers, and/or colonization/recolonization of the upper and lower river sections by different ancestral groups.
Genetic homogenization among different parts of the Tornio-Kalix River system is likely facilitated by the unusual bifurcation tributary (Tärendö River, K8, Fig. 1) connecting the two drainages about 180 to 250 km from the river mouths. It provides a potential route for adult salmon ascending the middle part of the Kalix to access Tornio headwaters, and vice versa. We observed no significant genetic differentiation between parr in the bifurcation and parr in most parts of the Kalix mainstem or the mid to lower Tornio (Table 2). This and the stronger IBD signal when using the route through the bifurcation as the shortest waterway, compared to not including it, support a hypothesis of gene flow via this route. Additionally, the bifurcation mixes water of the two rivers, potentially causing returning salmon navigating via olfactory cues (Petersson 2016) to stray between river mouths. Tagging studies have indeed found that around 7% of smolts originating from the Tornio River are recaptured as adults in the Kalix River (A. Romakkaniemi, unpublished data). This is however within typical Atlantic salmon straying rates between watersheds, estimated from other studies (3–10%, Stabell 1984; Jonsson et al. 2003; Keefer and Caudill 2014).
Cauwelier et al. (2018b) examined 11 Atlantic salmon rivers in Scotland and observed a similar pattern of upstream–downstream rather than among-river genetic structuring, associated with exceptionally high rates of straying between rivers (27.4%). This suggests an alternative hypothesis that factors independent of the bifurcation are maintaining the parallel upstream–downstream structure in the Kalix and Tornio. In particular, a genetic basis to timing of freshwater entry and/or migration duration in the river could cause strays originating from the upper section of one river to preferentially spawn in the upper reaches of the alternative river, thus mediating gene flow between these distant headwater sites.
Life history variation within the river system
Our results imply substantial life history variation among salmon spawning in different parts of the Tornio River system. Specifically, salmon with a higher P(Upper), that were more likely to originate from stretches higher up in the river, spent more years feeding in freshwater before smoltification, entered the marine environment later during their smolt migration, rarely matured after just one year at sea, and returned to freshwater at an earlier date, compared to fish with a lower P(Upper), that were more likely to originate from the downstream reaches. Such variation in Atlantic salmon life history timing is present across and within most river systems, and is known to have both an environmental and genetic component (e.g. Thorstad et al. 2011, Barson et al. 2015).
In line with our findings, many studies have found that adult salmon entering rivers early in the season tend to be older (e.g. Jokikokko et al. 2004; Quinn et al. 2006; Harvey et al. 2017) and originate from higher up in the systems than later-returning individuals (Shearer 1990; Økland et al. 2001; Stewart et al. 2002; Niemelä et al. 2006; Östergren 2006). Vähä et al. (2011a) studied 1SW salmon entering the Teno River, and found that individuals from subpopulations in smaller tributaries entered freshwater earlier. They proposed that the earlier ascending adults have an advantage in situations with competition for limited spawning sites. Ascending the river early may also allow large salmon to reach their spawning sites before water becomes too shallow for them in the course of the season (Niemelä et al. 2006). It is also possible that salmon returning to the upper reaches of the Tornio enter the river earlier simply because they have a longer way (up to 450 km) to swim to their spawning grounds than downstream salmon. Conversely, the long distance could be the reason for upstream smolts to reach the sea later than their downstream counterparts. However, if smolts throughout the river aim to reach the sea at the same optimal conditions and date, smolts from the upper reaches could be expected to initiate their migration earlier in the season (Stewart et al. 2006). It should also be noted that as only one 1SW salmon was sampled in 2010, it is difficult to make fully robust conclusions of age structure differences of returning adults between the reporting units.
Variation in seasonal run timing of adults in multiple populations is associated with genetic differences, including a possible locus of large effect (e.g. Stewart et al. 2002; Cauwelier et al. 2018a; Pritchard et al. 2018). Differences in smolt migration timing among populations has also been suggested to be adaptive and have a genetic basis (Stewart et al. 2006; Thorstad et al. 2012; Harvey et al. 2020). In addition, the age at smoltification seems to be an adaptive and highly heritable feature influenced by genetics (e.g. Páez et al. 2011; Pedersen et al. 2013), while also being controlled by environmental cues and conditions (e.g. Otero et al. 2014). For example, a shorter growing season and subsequently worse conditions for juvenile growth increase smolt age of Atlantic salmon (Metcalfe and Thorpe 1990). The uppermost sampling sites in the Tornio River system experience around 10% more days with ice cover than the lowermost sites (220 days vs. 200 days, Korhonen 2006), which could partially explain why smolts with higher P(Upper) were on average emigrating at an older age.
Implications for conservation and management
Currently, salmon in the Tornio and Kalix Rivers are managed as two separate stocks. Our analyses suggest that this may not be warranted from the conservation genetic perspective. Instead, an overall aim should be to retain the genetic and life history diversity present in both rivers, by ensuring the security of populations throughout these large river systems across Finland and Sweden. Therefore, our findings highlight the importance of cross-border cooperation in the management of these two salmon rivers. However, before a final recommendation on the most appropriate management strategy can be made, it would be recommendable to gain a clearer picture of possible genetic structure at potentially adaptive loci, and also assess how well evolutionary and ecological processes match in the system. As discussed for example by Waples and Gaggiotti (2006), ecological and evolutionary population concepts do not always go “hand in hand”, and in some situations a high rate of gene flow (evolutionary process) can still be accompanied by relatively low demographic exchange (ecological process). In the present case, straying between the lower Tornio and Kalix appears high enough to prevent genetic differentiation, but may still be low enough to allow largely independent demographic dynamics in the separate rivers. Hence additional research is warranted before recommending changes with respect to practical fisheries management (i.e. data collection and stock assessment).
Our results suggest that the salmon populations in the upper reaches of the Tornio-Kalix system harbour important genetic and life history diversity. These populations may be particularly vulnerable to the effects of harvesting both at sea and in the rivers due to their longer generation time and longer riverine migrations, increasing the chance of mortality before reproduction (e.g. Garant et al. 2003). Moreover, similar to Jansson (1993), we observed slightly lower allelic richness and mean heterozygosity in the upper than the lower sections of the Tornio-Kalix system. As genetic diversity is considered a prerequisite for evolutionary potential and population health, reduced genetic variation may indicate that the upstream populations in the Tornio-Kalix system are more vulnerable to environmental changes.
In the sea, Baltic salmon are harvested throughout their feeding and spawning migrations. Harvesting on the marine feeding grounds has markedly decreased over the last three decades, partly due to reduced catch quotas that have reduced overall sea fishing pressure (ICES 2019). This is expected to have increased survival of later maturing salmon in particular, as natural and fishery-related mortality at sea for Baltic salmon is high (ICES 2019), and may thus have benefitted salmon from the upper reaches of the Tornio and Kalix. Coastal fishing targets adult salmon migrating back to their natal river to spawn, and has also decreased due to quota restrictions. Additionally, delayed opening of the coastal fishing season of Baltic salmon since the 1980s is thought to have contributed to the recovery of the Tornio-Kalix salmon stock (Romakkaniemi et al. 2003). Salmon of the Tornio and Kalix are particularly targeted by fisheries along the Finnish coast (Whitlock et al. 2018). After two decades of delayed opening of the coastal fishing season, recent regulatory changes in the Finnish coastal fisheries now allow limited harvesting of salmon in the early season. Our results show that salmon entering the Tornio River early appear to be largely on the way to the upper river reaches, and allowing early fishing may thus increase fishing pressure on the upstream populations. Therefore, it is particularly important to closely follow and study what consequences changed temporal regulations of the coastal fishing may have on the Tornio and Kalix salmon populations.
In the Tornio River, salmon assigned to the ‘Upper’ reporting group are strongly over-represented in the adult samples caught by anglers, compared to the smolt samples caught in the smolt trap (29.4% of adults in 2009, 45.0% of adults in 2010; 6.6% of smolts). This observation could be driven by two phenomena relevant to management of the salmon stocks. First, upstream fish may have an increased rate of survival in the marine environment compared to downstream fish, for unknown reasons. Second, there may be disproportionate riverine fishing pressure on Tornio fish originating from the upstream reaches. Our adult sampling approximates total riverine fishery catches well along the main fishing area, where angling pressure and catches per km appear to be the highest: Finnish anglers catch a large majority of the total salmon catch in the Tornio River system, and about one-third of their total catch was caught from this 70 km long stretch in 2009–2010 (Vähä et al. 2010, 2011b; Palm et al. 2020). However, we caution that this observation may also be due to biased sampling, for five reasons. First, the samples were taken in different years (adults 2009–2010; smolts 2011). Second, while smolt samples were collected over the entire known emigration period, adult samples were restricted to the legal fishing season, which may not overlap the entire return migration period. Third, if the temporal window for migration of upstream smolts is much narrower than that of smolts from lower reaches, taking a maximum of 5 smolt samples per day would under-sample the former. Fourth, the adult fishing location was above the lowest Tornio sites. Finally, the distribution of our adult samples was mildly biased to the early fishing season, compared to the total catch. We recommend further investigation to confirm whether upstream adults are disproportionately targeted in the riverine fishery.
Run timing differences associated with different river sections makes it possible to manage specific stocks in river systems via temporal fishing closures (Vähä et al. 2011a; Cauwelier et al. 2018a), which could be utilized also in the local fishery management of the Tornio-Kalix. Such measures targeted on harvest timing have the potential to alter population trajectories and permit conservation or expansion of certain stock subcomponents (Kallio-Nyberg et al. 2011; Harvey et al. 2017; Erkinaro et al. 2019). Furthermore, variation in migration patterns is an important part of the genetic and phenotypic diversity of fish stocks, and thus essential for the resilience of the populations that fisheries depend on (Quinn et al. 2016; Jacobson et al. 2019; Tamario et al. 2019). Conserving the observed life history diversity could therefore be of large benefit for the long-term survival of the Tornio-Kalix salmon stock. To this end, understanding the evolutionary forces potentially promoting the upstream–downstream differentiation in the Tornio and Kalix Rivers would help in defining relevant management and conservation units in the system. Temporal studies incorporating genomic data from the river system could provide useful further information for the conservation of these wild salmon populations.
Availability of data and material (data transparency)
Data used in the analyses are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.kh1893245.
References
Anderson EC, Waples RS, Kalinowski ST (2008) An improved method for predicting the accuracy of genetic stock identification. Can J Fish Aquat Sci 65:1475–1486. https://doi.org/10.1139/F08-049
Anttila P, Romakkaniemi A, Kuusela J, Koski P (2008) Epidemiology of Gyrodactylus salaris (Monogenea) in the River Tornionjoki, a Baltic wild salmon river. J Fish Dis. https://doi.org/10.1111/j.1365-2761.2008.00916.x
Barson NJ, Aykanat T, Hindar K et al (2015) Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature 528:405–408. https://doi.org/10.1038/nature16062
Bourret V, Kent MP, Primmer CR et al (2013) SNP-array reveals genome-wide patterns of geographical and potential adaptive divergence across the natural range of Atlantic salmon (Salmo salar). Mol Ecol 22:532–551. https://doi.org/10.1111/mec.12003
Cairney M, Taggart JB, Høyheim B (2000) Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Mol Ecol 9:2155–2234. https://doi.org/10.1046/j.1365-294X.2000.01053.x
Cauwelier E, Gilbey J, Sampayo J et al (2018a) Identification of a single genomic region associated with seasonal river return timing in adult Scottish Atlantic salmon (Salmo salar), using a genome-wide association study. Can J Fish Aquat Sci 75:1427–1435. https://doi.org/10.1139/cjfas-2017-0293
Cauwelier E, Stewart DC, Millar CP et al (2018b) Across rather than between river genetic structure in Atlantic salmon Salmo salar in north-east Scotland, UK: potential causes and management implications. J Fish Biol 92:607–620. https://doi.org/10.1111/jfb.13542
Cullingham CI, Miller JM, Peery RM et al (2020) Confidently identifying the correct K value using the DeltaK method: when does K = 2? Mol Ecol 29:862–869. https://doi.org/10.1111/mec.15374
Dillane E, Cross MC, McGinnity P et al (2007) Spatial and temporal patterns in microsatellite DNA variation of wild Atlantic salmon, Salmo salar, in Irish rivers. Fish Manag Ecol 14:209–219. https://doi.org/10.1111/j.1365-2400.2007.00544.x
Dillane E, McGinnity P, Coughlan JP et al (2008) Demographics and landscape features determine intrariver population structure in Atlantic salmon (Salmo salar L.): the case of the River Moy in Ireland. Mol Ecol 17:4786–4800. https://doi.org/10.1111/j.1365-294X.2008.03939.x
Dionne M, Caron F, Dodson JJ, Bernatchez L (2009) Comparative survey of within-river genetic structure in Atlantic salmon; Relevance for management and conservation. Conserv Genet 10:869–879. https://doi.org/10.1007/s10592-008-9647-5
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Ensing D, Prodöhl PA, McGinnity P et al (2011) Complex pattern of genetic structuring in the Atlantic salmon (Salmo salar L.) of the River Foyle system in northwest Ireland: disentangling the evolutionary signal from population stochasticity. Ecol Evol 1:359–372. https://doi.org/10.1002/ece3.32
Erkinaro J, Czorlich Y, Orell P et al (2019) Life history variation across four decades in a diverse population complex of atlantic salmon in a large subarctic river. Can J Fish Aquat Sci 76:42–55. https://doi.org/10.1139/cjfas-2017-0343
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587. https://doi.org/10.1001/jama.1987.03400040069013
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1080/00837792.1905.10669550
Fleming IA (1996) Reproductive strategies of Atlantic salmon: ecology and evolution. Rev Fish Biol Fish 6:379–416. https://doi.org/10.1007/BF00164323
Fontaine PM, Dodson JJ, Bernatchez L, Slettan A (1997) A genetic test of metapopulation structure in Atlantic salmon (Salmo salar) using microsatellites. Can J Fish Aquat Sci 54:2434–2442. https://doi.org/10.1139/f97-152
Frantz AC, Cellina S, Krier A et al (2009) Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: clusters or isolation by distance? J Appl Ecol 46:493–505. https://doi.org/10.1111/j.1365-2664.2008.01606.x
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW (2012) Harnessing genomics for delineating conservation units. Trends Ecol Evol 27:489–496. https://doi.org/10.1016/j.tree.2012.05.012
Garant D, Dodson JJ, Bernatchez L (2000) Ecological determinants and temporal stability of the within-river population structure in Atlantic salmon (Salmo salar L.). Mol Ecol 9:615–628. https://doi.org/10.1046/j.1365-294X.2000.00909.x
Garant D, Dodson JJ, Bernatchez L (2003) Differential reproductive success and heritability of alternative reproductive tactics in wild Atlantic salmon (Salmo salar L.). Evolution 57:1133–1141
Gilbey J, Verspoor E, McLay A, Houlihan D (2004) A microsatellite linkage map for Atlantic salmon (Salmo salar). Anim Genet 35:98–105. https://doi.org/10.1111/j.1365-2052.2004.01091.x
Griffiths AM, Machado-Schiaffino G, Dillane E et al (2010) Genetic stock identification of Atlantic salmon (Salmo salar) populations in the southern part of the European range. BMC Genet. https://doi.org/10.1186/1471-2156-11-31
Hansen MM, Nielsen EE, Mensberg KLD (1997) The problem of sampling families rather than populations: relatedness among individuals in samples of juvenile brown trout Salmo trutta L. Mol Ecol 6:469–474. https://doi.org/10.1046/j.1365-294X.1997.t01-1-00202.x
Harvey AC, Tang Y, Wennevik V et al (2017) Timing is everything: fishing-season placement may represent the most important angling-induced evolutionary pressure on Atlantic salmon populations. Ecol Evol 7:7490–7502. https://doi.org/10.1002/ece3.3304
Harvey AC, Glover KA, Wennevik V, Skaala Ø (2020) Atlantic salmon and sea trout display synchronised smolt migration relative to linked environmental cues. Sci Rep 10:1–13. https://doi.org/10.1038/s41598-020-60588-0
Hilborn R, Quinn TP, Schindler DE, Rogers DE (2003) Biocomplexity and fisheries sustainability. Proc Natl Acad Sci USA 100:6564–6568. https://doi.org/10.1073/pnas.1037274100
Hutchinson WF (2008) The dangers of ignoring stock complexity in fishery management: the case of the North Sea cod. Biol Lett 4:693–695. https://doi.org/10.1890/03-5066
ICES (2011) Report of the workshop on age determination of Salmon (WKADS), 18–20 Jan 2011, Galway, Ireland. ICES CM 2011/ACOM:44
ICES (2019) Baltic Salmon and Trout Assessment Working Group (WGBAST). ICES Sci Rep 1:312. https://doi.org/10.17895/ices.pub.4979
Jacobson P, Gårdmark A, Huss M (2019) Population and size-specific distribution of Atlantic salmon Salmo salar in the Baltic Sea over 5 decades. J Fish Biol. https://doi.org/10.1111/jfb.14213
Janes JK, Miller JM, Dupuis JR et al (2017) The K = 2 conundrum. Mol Ecol 26:3594–3602. https://doi.org/10.1111/mec.14187
Jansson H (1993) Allozyme evidence for local populations of Atlantic salmon (Salmo salar L.) in the Kalix River, northern Sweden. ICES study group on stock identification protocols for finfish and shellfish stocks
Jokikokko E, Kallio-Nyberg I, Jutila E (2004) The timing, sex and age composition of the wild and reared Atlantic salmon ascending the Simojoki River, northern Finland. J Appl Ichthyol 20:37–42. https://doi.org/10.1111/j.1439-0426.2004.00491.x
Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour 10:551–555. https://doi.org/10.1111/j.1755-0998.2009.02787.x
Jonsson B, Jonsson N, Hansen LP (2003) Atlantic salmon straying from the River Imsa. J Fish Biol 62:641–657. https://doi.org/10.1046/j.1095-8649.2003.00053.x
Kallio-Nyberg I, Saloniemi I, Jutila E, Jokikokko E (2011) Effect of hatchery rearing and environmental factors on the survival, growth and migration of Atlantic salmon in the Baltic Sea. Fish Res 109:285–294. https://doi.org/10.1016/j.fishres.2011.02.015
Karlsson L, Karlström Ö (1994) The Baltic salmon (Salmo salar L.): its history, present situation and future. Dana 10:61–85
Karvonen A, Seehausen O (2012) The role of parasitism in adaptive radiations—when might parasites promote and when might they constrain ecological speciation? Int J Ecol. https://doi.org/10.1155/2012/280169
Keefer ML, Caudill CC (2014) Homing and straying by anadromous salmonids: a review of mechanisms and rates. Rev Fish Biol Fish 24:333–368. https://doi.org/10.1007/s11160-013-9334-6
King TL, Eackles MS, Letcher BH (2005) Microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure, and mixed-fishery analyses. Mol Ecol Notes 5:130–132. https://doi.org/10.1111/j.1471-8286.2005.00860.x
Koljonen M-L (2001) Conservation goals and fisheries management units for Atlantic salmon in the Baltic Sea area. J Fish Biol 59:269–288. https://doi.org/10.1006/jfbi.2001.1757
Koljonen M-L (2006) Annual changes in the proportions of wild and hatchery Atlantic salmon (Salmo salar) caught in the Baltic Sea. ICES J Mar Sci 63:1274–1285. https://doi.org/10.1016/j.icesjms.2006.04.010
Koljonen M-L, McKinnell S (1996) Assessing seasonal changes in stock composition of Atlantic salmon catches in the Baltic Sea with genetic stock identification. J Fish Biol 49:998–1018. https://doi.org/10.1006/jfbi.1996.0228
Koljonen M-L, Jansson H, Paaver T et al (1999) Phylogeographic lineages and differentiation pattern of Atlantic salmon (Salmo salar) in the Baltic Sea with management implications. Can J Fish Aquat Sci 56:1766–1780. https://doi.org/10.1139/f99-104
Kopelman NM, Mayzel J, Jakobsson M et al (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Korhonen J (2006) Long-term changes in like ice cover in Finland. Nord Hydrol 37:347–363. https://doi.org/10.2166/nh.2006.019
Kuusela AJ, Holopainen R, Meinilä M et al (2009) Clonal structure of Salmon parasite Gyrodactylus salaris on a coevolutionary gradient on Fennoscandian Salmon (Salmo salar). Ann Zool Fennici 46:21–33
Landry C, Bernatchez L (2001) Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar). Mol Ecol 10:2525–2539. https://doi.org/10.1046/j.1365-294X.2001.01383.x
Lumme J, Anttila P, Rintamäki P et al (2016) Genetic gradient of a host–parasite pair along a river persisted ten years against physical mobility: baltic Salmo salar vs. Gyrodactylus salaris. Infect Genet Evol 45:33–39. https://doi.org/10.1016/j.meegid.2016.08.006
McConnell SK, O’Reilly P, Hamilton L et al (1995) Polymorphic microsatellite loci from Atlantic salmon (Salmo salar): genetic differentiation of North American and European populations. Can J Fish Aquat Sci 52:1863–1872. https://doi.org/10.1139/f95-779
McConnell SKJ, Ruzzante DE, O’Reilly PT et al (1997) Microsatellite loci reveal highly significant genetic differentiation among Atlantic salmon (Salmo salar L.) stocks from the east coast of Canada. Mol Ecol 6:1075–1089. https://doi.org/10.1046/j.1365-294X.1997.00282.x
Metcalfe NB, Thorpe JE (1990) Determinants of geographical variation in the age of seaward-migrating Salmon, Salmo salar. J Anim Ecol 59:135–145
Moore JS, Bourret V, Dionne M et al (2014) Conservation genomics of anadromous Atlantic salmon across its North American range: outlier loci identify the same patterns of population structure as neutral loci. Mol Ecol 23:5680–5697. https://doi.org/10.1111/mec.12972
Moran BM, Anderson EC (2019) Bayesian inference from the conditional genetic stock identification model. Can J Fish Aquat Sci 76:551–560. https://doi.org/10.1139/cjfas-2018-0016
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19:153–170
Niemelä E, Orell P, Erkinaro J et al (2006) Previously spawned Atlantic salmon ascend a large subarctic river earlier than their maiden counterparts. J Fish Biol 69:1151–1163. https://doi.org/10.1111/j.1095-8649.2006.01190.x
Nilsson J, Gross R, Asplund T et al (2001) Matrilinear phylogeography of Atlantic salmon (Salmo salar L.) in Europe and postglacial colonization of the Baltic Sea area. Mol Ecol 10:89–102. https://doi.org/10.1046/j.1365-294X.2001.01168.x
Nordahl O, Koch-Schmidt P, Sunde J et al (2019) Genetic differentiation between and within ecotypes of pike (Esox lucius) in the Baltic Sea. Aquat Conserv Mar Freshw Ecosyst. https://doi.org/10.1002/aqc.3196
O’Reilly PT, Hamilton LC, McConnell SK, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci 53:2292–2298. https://doi.org/10.1139/f96-192
Økland F, Erkinaro J, Moen K et al (2001) Return migration of Atlantic salmon in the River Tana: phases of migratory behaviour. J Fish Biol 59:862–874. https://doi.org/10.1006/jfbi.2001.1701
Östergren J (2006) Migration and Genetic Structure of Salmo salar and Salmo trutta in Northern Swedish Rivers. Dissertation, Swedish University of Agricultural Sciences
Östergren J, Palm S, Gilbey J, Dannewitz J (2020) Close relatives in population samples: evaluation of the consequences for genetic stock identification. Mol Ecol Resour 20:498–510. https://doi.org/10.1111/1755-0998.13131
Östman Ö, Olsson J, Dannewitz J et al (2017) Inferring spatial structure from population genetics and spatial synchrony in demography of Baltic Sea fishes: implications for management. Fish Fish 18:324–339. https://doi.org/10.1111/faf.12182
Otero J, L’Abée-Lund JH, Castro-Santos T et al (2014) Basin-scale phenology and effects of climate variability on global timing of initial seaward migration of Atlantic salmon (Salmo salar). Glob Chang Biol 20:61–75. https://doi.org/10.1111/gcb.12363
Ozerov MY, Veselov AE, Lumme J, Primmer CR (2013) Temporal variation of genetic composition in Atlantic salmon populations from the Western White Sea Basin: influence of anthropogenic factors? BMC Genet. https://doi.org/10.1186/1471-2156-14-88
Páez DJ, Brisson-Bonenfant C, Rossignol O et al (2011) Alternative developmental pathways and the propensity to migrate: a case study in the Atlantic salmon. J Evol Biol 24:245–255. https://doi.org/10.1111/j.1420-9101.2010.02159.x
Palm S, Romakkaniemi A, Dannewitz J et al (2020) Tornionjoen lohi-, meritaimen- ja vaellussiikakannat – yhteinen ruotsalais-suomalainen biologinen selvitys sopivien kalastussääntöjen arvioimiseksi vuodelle 2019 (In Finnish and Swedish)
Palsbøll PJ, Bérubé M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16. https://doi.org/10.1016/j.tree.2006.09.003
Paterson S, Piertney SB, Knox D et al (2004) Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol Ecol Notes 4:160–162. https://doi.org/10.1111/j.1471-8286.2004.00598.x
Pedersen S, Berg PR, Culling M et al (2013) Quantitative trait loci for precocious parr maturation, early smoltification, and adult maturation in double-backcrossed trans-Atlantic salmon (Salmo salar). Aquaculture 410–411:164–171. https://doi.org/10.1016/j.aquaculture.2013.06.039
Perez MF, Franco FF, Bombonato JR et al (2018) Assessing population structure in the face of isolation by distance: are we neglecting the problem? Divers Distrib 24:1883–1889. https://doi.org/10.1111/ddi.12816
Petersson E (2016) Homing and timing of reproduction. In: Vladic T, Petersson E (eds) Evolutionary biology of the Atlantic Salmon. CRC Press, Boca Raton, pp 44–59
Potter ECE, MacLean JC, Wyatt RJ, Campbell RNB (2003) Managing the exploitation of migratory salmonids. Fish Res 62:127–142. https://doi.org/10.1016/S0165-7836(02)00159-5
Primmer CR, Veselov AJ, Zubchenko A et al (2006) Isolation by distance within a river system: genetic population structuring of Atlantic salmon, Salmo salar, in tributaries of the Varzuga River in northwest Russia. Mol Ecol 15:653–666. https://doi.org/10.1111/j.1365-294X.2005.02844.x
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard VL, Mäkinen H, Vähä J-P et al (2018) Genomic signatures of fine-scale local selection in Atlantic salmon suggest involvement of sexual maturation, energy homeostasis and immune defence-related genes. Mol Ecol. https://doi.org/10.1111/mec.14705
Pruuki V (1993) Changes in the status of the salmon stock in the River Tornionjoki. ICES CM 1993/M: 51. + Figures
Quinn TP, McGinnity P, Cross TF (2006) Long-term declines in body size and shifts in run timing of Atlantic salmon in Ireland. J Fish Biol. https://doi.org/10.1111/j.1095-8649.2006.01017.x
Quinn TP, McGinnity P, Reed TE (2016) The paradox of “premature migration” by adult anadromous salmonid fishes: patterns and hypotheses. Can J Fish Aquat Sci 73:1015–1030. https://doi.org/10.1139/cjfas-2015-0345
Rambaut A (2012) FigTree v. 1.4.3. Available at http://tree.bio.ed.ac.uk/software/figtree/
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rice WR (1989) Analyzing tables of statistical tests. Am Sociol Rev 43:223–225. https://doi.org/10.1111/j.0014-3820.2001.tb00731.x
Romakkaniemi A (2008) Conservation of Atlantic salmon by supplementary stocking of juvenile fish. Dissertation, Helsinki University Print
Romakkaniemi A, Perä I, Karlsson L et al (2003) Development of wild Atlantic salmon stocks in the rivers of the northern Baltic Sea in response to management measures. ICES J Mar Sci 60:329–342. https://doi.org/10.1016/S1054-3139(03)00020-1
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106. https://doi.org/10.1111/j.1471-8286.2007.01931.x
Säisä M, Koljonen M-L, Gross R et al (2005) Population genetic structure and postglacial colonization of Atlantic salmon (Salmo salar) in the Baltic Sea area based on microsatellite DNA variation. Can J Fish Aquat Sci 62:1887–1904. https://doi.org/10.1139/f05-094
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Scheer BT (1939) Homing instinct in Salmon. Q Rev Biol 14:408–430
Schindler DE, Hilborn R, Chasco B et al (2010) Population diversity and the portfolio effect in an exploited species. Nature 465:609–612. https://doi.org/10.1038/nature09060
Shearer WM (1990) The Atlantic salmon (Salmo salar L.) of the North Esk with particular reference to the relationship between both river and sea age and time of return to home waters. Fish Res 10:93–123. https://doi.org/10.1016/0165-7836(90)90017-P
Slettan A, Olsaker I, Lie Ø (1995) Atlantic salmon, Salmo salar, microsatellites at the SSOSL25, SSOSL85, SSOSL311, SSOSL417 loci. Anim Genet 26:277–285
Spidle AP, Bane Schill W, Lubinski BA, King TL (2001) Fine-scale population structure in Atlantic salmon from Maine’s Penobscot River drainage. Conserv Genet 2:11–24. https://doi.org/10.1023/A:1011580217381
Spidle AP, Kalinowski ST, Lubinski BA et al (2003) Population structure of Atlantic salmon in maine with reference to populations from Atlantic Canada. Trans Am Fish Soc 132:196–209
Stabell OB (1984) Homing and olfaction in salmonids: a critical review with special reference to the Atlantic salmon. Biol Rev 59:333–388
Ståhl G (1981) Genetic differentiation among natural populations of Atlantic salmon (Salmo salar) in northern Sweden. Ecol Bull 95–105
Ståhl G (1987) Genetic population structure of Atlantic salmon. In: Ryman N, Utter F (eds) Population genetics and fisheries management. University of Washington Press, Seattle, pp 121–140
Stewart DC, Smith GW, Youngson AF (2002) Tributary-specific variation in timing of return of adult Atlantic salmon (Salmo salar) to fresh water has a genetic component. Can J Fish Aquat Sci 59:276–281. https://doi.org/10.1139/f02-011
Stewart DC, Middlemas SJ, Youngson AF (2006) Population structuring in Atlantic salmon (Salmo salar): evidence of genetic influence on the timing of smolt migration in sub-catchment stocks. Ecol Freshw Fish 15:552–558. https://doi.org/10.1111/j.1600-0633.2006.00197.x
Takezaki N, Nei M, Tamura K (2014) POPTREEW: web version of POPTREE for constructing population trees from allele frequency data and computing some other quantities. Mol Biol Evol 31:1622–1624. https://doi.org/10.1093/molbev/msu093
Tamario C, Sunde J, Petersson E et al (2019) Ecological and evolutionary consequences of environmental change and management actions for migrating fish. Front Ecol Evol 7:1–24. https://doi.org/10.3389/fevo.2019.00271
Thorstad EB, Whoriskey F, Rikardsen AH, Aarestrup K (2011) Aquatic nomads: the life and migrations of the Atlantic salmon. In: Aas Ø, Einum S, Klemetsen A, Skurdal J (eds) Atlantic salmon ecology, 1st edn. Blackwell Publishing Ltd., Oxford, pp 31–62
Thorstad EB, Whoriskey F, Uglem I et al (2012) A critical life stage of the Atlantic salmon Salmo salar: behaviour and survival during the smolt and initial post-smolt migration. J Fish Biol 81:500–542. https://doi.org/10.1111/j.1095-8649.2012.03370.x
Tonteri A, Koskinen MT, Lesbarrères D et al (2005) Phylogeography of anadromous and non-anadromous Atlantic salmon (Salmo salar) from northern Europe. Ann Zool Fennici 42:1–22
Vähä J-P, Erkinaro J, Niemelä E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol. https://doi.org/10.1111/j.1365-294X.2007.03329.x
Vähä J-P, Erkinaro J, Niemelä E, Primmer CR (2008) Temporally stable genetic structure and low migration in an Atlantic salmon population complex: implications for conservation and management. Evol Appl. https://doi.org/10.1111/j.1752-4571.2007.00007.x
Vähä V, Romakkaniemi A, Ankkuriniemi M et al (2010) Lohi- ja meritaimenkantojen seuranta Tornionjoen vesistössä vuonna 2009. (In Finnish, abstract in English)
Vähä JP, Erkinaro J, Niemelä E et al (2011a) Temporally stable population-specific differences in run timing of one-sea-winter Atlantic salmon returning to a large river system. Evol Appl. https://doi.org/10.1111/j.1752-4571.2010.00131.x
Vähä V, Romakkaniemi A, Ankkuriniemi M, et al (2011b) Lohi- ja meritaimenkantojen seuranta Tornionjoen vesistössä vuonna 2010 (In Finnish, abstract in English)
Vähä J-P, Erkinaro J, Falkegård M et al (2017) Genetic stock identification of Atlantic salmon and its evaluation in a large population complex. Can J Fish Aquat Sci. https://doi.org/10.1139/cjfas-2015-0606
Venables WN, Ripley BD (2002) Modern applied statistics with S, 4th edn. Springer, New York
Verspoor E, Beardmore JA, Consuegra S et al (2005) Population structure in the Atlantic salmon: insights from 40 years of research into genetic protein variation. J Fish Biol 67:3–54. https://doi.org/10.1111/j.1095-8649.2005.00838.x
Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Wang J (2004) Sibship reconstruction from genetic data with typing errors. Genetics 166:1963–1979. https://doi.org/10.1534/genetics.166.4.1963
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439. https://doi.org/10.1111/j.1365-294X.2006.02890.x
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358. https://doi.org/10.2307/2408641
Wellband K, Mérot C, Linnansaari T et al (2018) Chromosomal fusion and life history-associated genomic variation contribute to within-river local adaptation of Atlantic salmon. Mol Ecol. https://doi.org/10.1111/mec.14965
Whitlock R, Mäntyniemi S, Palm S et al (2018) Integrating genetic analysis of mixed populations with a spatially explicit population dynamics model. Methods Ecol Evol 2018:1017–1035. https://doi.org/10.1111/2041-210X.12946
Acknowledgements
The study was funded by fishing license revenues from the Tornio/Torne River, The Swedish Agency for Marine and Water Management, Grant/Award 2013-1288 to JÖ from the Swedish Research Council Formas and grants to AM from Societas pro Fauna et Flora Fennica, The Betty Väänänen Fund from The Kuopio Naturalists’ Society (KLYY), The Raija and Ossi Tuuliainen Foundation, and The Finnish Foundation for Nature Conservation’s Baltic Sea Fund. We would like to thank Anders Asp (original map used for Fig. 1), field workers and other staff for assistance with the research material (Markku Kilpala, Stefan Stridsman, Ville Vähä, Kari Pulkkinen, Matti Ankkuriniemi, Irmeli Torvi and Rauno Hokki), Tore Prestegaard and Linda Söderberg for lab assistance, Paul Debes for statistical discussions, two anonymous reviewers for their helpful comments that improved the manuscript, and the anglers who collected the adult samples along the Tornio River.
Funding
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. The study was funded by fishing license revenues from the Tornio River, The Swedish Agency for Marine and Water Management, The Swedish Research Council Formas (Grant/Award 2013-1288 to JÖ), Societas pro Fauna et Flora Fennica, The Betty Väänänen Fund from The Kuopio Naturalists’ Society (KLYY), The Raija and Ossi Tuuliainen Foundation (Raija ja Ossi Tuuliaisen Säätiö), and The Baltic Sea Fund from The Finnish Foundation for Nature Conservation (Suomen Luonnonsuojelun Säätiö) (grants to AM).
Author information
Authors and Affiliations
Contributions
Project design: JD, SP, AR, JÖ; Sample collection: AR, JÖ; Laboratory work: EL; Data analysis: JD, AM, SP, VLP, JÖ; Paper writing: AM, SP, VLP, AR; Other resources: CRP; Commenting on manuscript: All authors.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Consent for publication
All authors consent to submitting this article to Conservation Genetics.
Code availability (software application or custom code)
R codes used in the data analyses are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.kh1893245.
Ethical approval
This study used Atlantic salmon tissue samples that had been collected as part of ongoing fisheries monitoring programs. All sample collection complied with applicable national legislation (Finland or Sweden).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miettinen, A., Palm, S., Dannewitz, J. et al. A large wild salmon stock shows genetic and life history differentiation within, but not between, rivers. Conserv Genet 22, 35–51 (2021). https://doi.org/10.1007/s10592-020-01317-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01317-y