Abstract
Management and restoration planning for Pacific salmon is often characterized by efforts at broad multi-basin scales. However, finer-scale genetic and phenotypic variability may be present within individual basins and can be overlooked in such efforts, even though it may be a critical component for long-term viability. Here, we investigate Chinook salmon (Oncorhynchus tshawytscha) within the Siletz River, a small coastal watershed in Oregon, USA. Adult Chinook salmon were genotyped using neutral microsatellite markers, single nucleotide polymorphisms and “adaptive” loci, associated with temporal variation in migratory behavior in many salmon populations, to investigate genetic diversity based upon both spatial and temporal variation in migratory and reproductive behavior. Results from all three marker types identified two genetically distinct populations in the basin, corresponding to early returning fish that spawn above a waterfall, a spring-run population, and later returning fish spawning below the waterfall, a fall-run population. This finding is an important consideration for management of the species, as spring-run populations generally only have been recognized in large watersheds, and highlights the need to evaluate population structure of salmon within smaller watersheds, and thereby increase the probability of successful conservation of salmon species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic and life-history diversity contribute to the resilience of native species in dynamic environments. Chinook salmon (Oncorhynchus tshawytscha) in western North America historically exhibited at least four behavioral life histories that are associated with the season of adult upstream migration (fall, spring, winter, and summer runs) and maturation status (Yoshiyama et al. 1998). Contributing to evolution of these migratory patterns is their homing fidelity to natal spawning rivers that allow for reproductive isolation and development of unique evolutionary trajectories (Waples 2001; Quinn 2004). Anthropogenic activities including harvest, waterway development, hatchery production, and land use practices have altered salmon populations and their associated freshwater ecosystems; the result is often reduced life-history and genetic variability (Yoshiyama et al. 1998; Brenkman et al. 2012).
To counter the effects of declining populations, substantial focus has been directed toward increasing abundance (supplementation programs) while also maintaining genetic diversity within evolutionarily significant units (ESUs) or other management units (Waples 1998; Olsen et al. 2000; Lindley et al. 2004; Eldridge and Killebrew 2007). As genetic techniques have improved, they have been used increasingly to enable better management (Clemento et al. 2014; Shafer et al. 2015). Salmonid fishes were among the first groups of organisms to be studied with molecular techniques, when the available methods were limited to the evaluation of genetically determined variation in a handful of proteins that could be reliably stained for detection (Utter and Hodgins 1972). One important application of molecular techniques has been genetic stock identification (GSI), which uses a “baseline” reference dataset of genotypes from individuals of known origin to identify the most likely provenance of individuals of unknown origin on the basis of their genotype (Milner et al. 1985). A North America-wide baseline dataset has been developed for Chinook salmon that includes 42 major reporting units (Seeb et al. 2007) and, more recently, baseline datasets using single nucleotide polymorphisms (SNPs) have been developed for regional applications, including in the California Current ecosystem (Clemento et al. 2014).
Pacific salmon management and restoration planning is characterized by efforts at broad multi-basin scales; however, fine-scale genetic structure that may be present within small watersheds (< 120 river km) is often overlooked in recovery planning. Homing to natal stream of origin is an influential driver of isolation among spawning groups and contributes to the genetic structure that has been observed at the landscape scale within salmon species, as do other ecological factors (i.e., habitat fragmentation, water development, episodic landslides or fires etc.). However, proper management of this fine-scale life history variability is a critical component in the long-term population persistence in a dynamic landscape.
In addition to geographically structured genetic variation, salmon species demonstrate substantial life-history variation (for detailed descriptions of life history variation see (Groot and Margolis 1991; Waples 2001). In Chinook salmon, much of this variation manifests as run-timing “ecotypes” which are characterized by mean date of freshwater entry and reproductive status (Waples et al. 2004; Moran et al. 2013). The ‘fall-run’, which enter freshwater as reproductively mature adults in late summer and fall, is regionally dominant in the southern extreme of the species range in North America. The ‘spring-run’ ecotype, which enter freshwater as reproductively immature adults in spring and early summer and hold in deep pools before maturing in the fall, are much less abundant and are derived from proximate fall-run populations in coastal basins (Kinziger et al. 2013).
Here, we investigated if fine-scale population structure of Chinook salmon exists in the Siletz River, a small watershed in coastal Oregon. Previous studies that examined salmon populations from the Siletz River using data from neutral genetic variation included only samples from the fall-run, and were part of large-scale efforts to characterize genetic structure in the species and provide methods for identification of fish of unknown origin (Seeb et al. 2007; Moran et al. 2013; Clemento et al. 2014). We studied salmon of different life-history types and used data from three types of molecular marker to: i) identify patterns of neutral genetic variation within the watershed, ii) evaluate whether temporal patterns of life-history variation are correlated with variation at genetic markers known to be associated with run-timing, and iii) determine phylogeographic patterns within the river and compare them to other coastal basins in the North Oregon Coast region. We then address the implications of our results for current conservation and management activities.
Methods
Study area
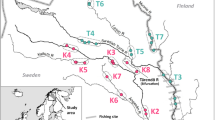
The Siletz River system (~109 river km) has headwaters in the Central Oregon Coast Range and meets the ocean at Siletz Bay. The geology of the watershed is principally volcanic. A waterfall that may be a barrier to upstream migration by aquatic organisms is located at river kilometer 103.8 (Fig. 1). A fish ladder was constructed in 1953 to allow fish passage during the winter for migratory fishes and Oregon Department of Fish and Wildlife (ODFW) operates it year round to control fish passage. In 1994, management practices began limiting access of anadromous fish that compete for spawning habitat with wild summer steelhead (Siletz basin fish management plan; ODFW 1997). Substantial changes to water flow and suitable salmon spawning habitat also occurred as a result of historical logging and splash-damming and alteration to the floodplains.
Map of sample sites for Chinook salmon from the Siletz River basin. Sampling locations are identified numerically. Each dot represents the downstream reach identification for ODFW sampling locations within Siletz River. Numbered sites indicate locations where genetic samples were collected for analysis. Sites 3, 4, 9, 13, 23 and 24 are located around river confluences; each inlay expands the area at these sites
As many as three Chinook salmon ecotypes may exist in the basin: spring-, summer-, and fall-run (Stan van de Wetering, Siletz Tribe, pers. comm). The largest of these is the fall-run, which enters the river in September. The spring-run enters the river in May and moves the farthest upstream; while the summer-run ecotype returns in July and spawns between mid-September and mid-October (Stan van de Wetering, Siletz Tribe, pers. comm.).
Sample collection
Tissue samples from Chinook salmon were collected by ODFW during their yearly carcass surveys and through monitoring at the trap on the fish ladder (Fig. 1). Samples that were collected during the fall carcass survey consisted of a small section of the least degraded flesh from each carcass. Body condition and geographic (reach ID) location was recorded for all samples. Collections occurred September through December in 2011 and 2012 and were considered to be from the fall-run ecotype (SIFA). These samples were collected downstream of the fish trap (Reach IDs 2–23; Fig. 1) and a total of 565 samples of sufficient quality for successful genotyping were collected.
Samples from carcasses of fish that appeared in the main stem Siletz River prior to October 15th in 2012 and 2013 were hypothesized to be a unique group, based on adult return and spawn time and were designated lower river early (SILE) fish. These samples were collected downstream at sampling locations Reach IDs 17–21 (Fig. 1). There were 55 such samples of sufficient quality for successful genotyping
The fish ladder and trap are operated year round, and enable the upstream passage of all early returning Chinook salmon. Fish that entered the trap were considered to be the spring-run ecotype (SISP) and were passed over the waterfall and allowed access to spawning grounds located upstream (Fig. 1). Scales were removed for age analysis and genotyping from all passed fish for which it was possible (i.e. body condition). During the sampling years, 700 fish were passed upstream at the fish trap and, of these, 258 individuals were sampled and genotyped.
Genetic analysis
Total genomic DNA was isolated from each tissue sample following the extraction protocol of Ivanova et al. (2006). Samples were genotyped with up to three types of molecular marker: presumably neutral microsatellites, putatively adaptive microsatellites, associated with run-timing variation, and single nucleotide polymorphisms (SNPs). Neutral microsatellite markers are not known to be associated with phenotypic expression and are considered to be selectively neutral; therefore, we were able to infer demographic processes that shaped population structure. Polymerase chain reaction (PCR) was used to amplify 21 neutral microsatellites following published thermocycling protocols. These included 10 microsatellites that are standardized range-wide (Moran et al. 2013) and an additional 11 microsatellites (Nelson and Beacham 1998; Naish and Park 2002; Williamson et al. 2002). The standardized microsatellite panel was developed for range-wide genetic stock identification (GSI) of Chinook salmon and included genotypes from populations throughout North America (Seeb et al. 2007; Moran et al. 2013). Fluorescent PCR products were electrophoresed on a 96-capillary DNA sequencer (3730XL; Applied Biosystems Inc.) and genotypes called using genemapper v3.7 software (Applied Biosystems).
Putatively adaptive microsatellite markers are associated with phenotypic expression and may be useful for interpreting adaptation of individuals to their local environments. The circadian clock gene network has been identified in salmon as contributing to the genetic control of adult migration timing in salmon (O'Malley et al. 2007, 2010). Three circadian clock gene markers, Ots515NWFSC (Ots515), Cryptochrome3 (Cry3), and OtsClock1b (clock1b), were amplified via published PCR and thermal cycling protocols. Ots515 is a QTL-linked marker that is associated with spawn time and body weight in rainbow trout (O'Malley et al. 2007). Cry3 is linked to flavoproteins that mediate circadian rhythms in plants (O'Malley et al. 2010). Clock1b contains a polyglutamine repeat tract that has been shown to vary in Chinook salmon (O'Malley et al. 2010).
Single nucleotide polymorphisms (SNPs) are sites in the genome that have two nucleotides segregating and are ubiquitous in vertebrates. A panel of 96 SNPs specific for Chinook salmon (Clemento et al. 2014) were genotyped on 96.96 Dynamic Arrays with an EP1 System (Fluidigm Corp., South San Francisco, CA), and genotypes were called with Fluidigm Genotyping Analysis software v2.1.1. These SNPs have been used previously to construct a GSI “baseline” database that is comprised of genotypes from more than 69 populations, including fall-run from the Siletz River. A total of 188 samples, 94 fall-run and 94 spring-run, chosen representatively from the larger sets of samples was genotyped with these markers to determine phylogenetic relationships among proximate coastal basins.
Statistical analysis of genetic variation
Loci were assessed for genotyping problems including null alleles or allelic dropout using microchecker (Van Oosterhout et al. 2004). Observed allele frequencies were tested for evidence of deviations from Hardy-Weinberg expectations (HWE) and for significant linkage disequilibrium (LD) between loci with genetix (Belkhir et al. 1996–2004). Summary statistics of genetic diversity were calculated for each hypothesized population. Characterization of the genetic diversity among loci was evaluated using allelic richness (Ar) a measure of allelic number that corrects for unequal sample sizes using a rarefaction method with hp_rare (Kalinowski 2005). The number of alleles (A), and observed (Ho) and expected (He) heterozygosity were calculated in genalex (Peakall and Smouse 2012).
Spatial structure was evaluated in three ways: pairwise estimates of the fixation index (FST), exact tests for genic and genotypic frequencies, and individual assignment tests. Pairwise FST values (θ; Weir and Cockerham 1984) were calculated in the program genalex and the data set was permuted 1000 times to determine if the values differed significantly from zero, an indication that populations may be genetically distinct. Exact tests for differences in genic and genotypic frequencies were performed with the program genepop (Raymond and Rousset 1995), which applies a Markov Chain Monte Carlo algorithm to account for small sample sizes or low-frequency alleles. Significant values of genic and genotypic frequencies may occur even though sufficient power to detect genetic differentiation through other methods is not possible, as is the case with populations where high gene flow exists.
A model-based Bayesian clustering method was used as an additional method to identify the degree of differentiation between the hypothesized populations. The software package structure v2.2 (Pritchard et al. 2000) estimates the likelihood for hypothesized values of k, the number of genetically distinct clusters or populations from which the sampled individuals were drawn. This method allows the data to define the clusters and assigns individuals to the k clusters without a priori information about their sampling locations. Five independent runs were performed for each value of k (2–6), using 50,000 burn-in and 150,000 retained iterations. An additional STRUCTURE run to assess the association of SISP and SIFA to the Central Oregon Coast reporting unit was conducted using published data provided in (Clemento et al. 2014). Five independent runs were also performed for values of k = 4–10, using the same burn-in and iterations as above.
Phylogeography
Phylogeographic patterns of fish within the Siletz River were inferred with a dendrogram based on SNP genotypes, constructed using chord distances (Cavalli-Sforza and Edwards 1967) and with the topology determined using the neighbor-joining algorithm, in the phylip package (Felsenstein 1993). Majority-rule consensus values were calculated from 1000 bootstrap replicates of the data by the phylip components seqboot and consense. Only bootstrap values above 80% were reported.
Results
Neutral genetic variation
Null alleles or other problems were identified in three microsatellite loci (Ots209, Ots211, and Omm1080), due to the presence of more homozygous individuals than would be expected. Significant departures from HWE existed for seven markers (Ots9, Ots104, OtsG409, Ogo4, Ots208, Ots249, and OtsG83) after adjusting for multiple comparisons, the remaining 11 loci were used for analysis. Allelic richness and heterozygosity of SISP was greater than that of either SIFA or SILE (Table 1). Linkage disequilibrium was found in all populations; the largest fraction of locus pairs in LD was within SISP (Table 1). Overall accuracy of individual assignment to population of origin using the microsatellite data was greater than 83% (Table 2). Pairwise FST values across years (2011 and 2012) were low (SIFA, FST = 0.001, p = 0.006; SISP, FST = 0.003, p = 0.001) and marginally significantly different from zero, but did not likely represent biologically meaningful differentiation (Hedrick 1999). Data were therefore pooled across years within groups for subsequent analyses.
Of the 96 SNP loci, four loci (Ots_108735–302, Ots_118175–479, Ots_128302–57, and Ots_Pr12) did not yield genotypes within the SISP population, and OkiOts_120255 functions to discriminate Chinook salmon from closely related coho salmon (O. kisutch); the remaining 91 loci were used for subsequent analyses (suppl. 1). There were no departures from HWE among loci following correction for multiple comparisons. Allelic richness and heterozygosity of SISP was again greater than that of either SIFA or SILE (Table 1). There was no evidence of linkage disequilibrium within SIFA; however, a small amount of LD was present in the SISP group (Table 1). The SNP dataset had similar ability to accurately assign individuals to population of origin as the microsatellite dataset (accuracy >85%; Table 2).
Pairwise FST differed significantly from zero between SILE and SISP, but not between SIFA and SILE (Table 3). Model-based clustering analysis with structure provided evidence of two major genetic groups in the Siletz River that corresponded to the fall-run and spring-run Chinook salmon ecotypes (Fig. 2a and b) and was consistent with the pattern identified by other analyses with both datasets.
Fractional ancestry plots obtained from STRUCTURE for a) microsatellite markers, b) SNPs, and c) SNPs using Clemento et al. (2014) genotypes for the North Oregon Coast Reporting Unit. Each vertical bar represents an individual’s genotype and the probability of being assigned to one of k (k = 2 or 6) genetically distinct clusters
Temporal adaptive genetic variation
Variation at the three markers associated with circadian clock genes provided further evidence for differentiation between migratory ecotypes of Chinook salmon in the Siletz River. All pairwise FST values and exact tests of genic and genotypic divergence between SISP and both SIFA and SILE, except those for locus Clock1b, were significant, indicating differentiation of SISP and both SIFA and SILE (Table 4). In contrast, these loci provided minimal evidence for differentiation between the earlier returning (SILE) and later returning lower river fish (SIFA), with non-significant FST values and significant tests of genic and genotypic differentiation at loci OTS515 only (Table 4).
Phylogeography
A neighbor-joining dendrogram was created with the SNP dataset and was compared to a larger published study of Chinook salmon by Clemento et al. (2014), that used the same loci and in which “reporting groups” of populations from the same geographical regions were identified. Siletz River fall-run Chinook salmon were grouped with the North Oregon Coast reporting unit in that study, as they were in several other genetic studies (Narum et al. 2008; Moran et al. 2013). Our analysis also placed SIFA in the North Oregon Coast reporting unit (bootstrap 83%; Fig. 3), which is consistent with the fact that the SIFA samples were collected from the same general location as the fall-run Siletz River fish that were analyzed in these previous studies. The Siletz River spring-run ecotype (SISP) population also branched with the North Oregon Coast reporting unit. Model-based clustering analysis with structure also supports this finding (Fig. 2c).
Mid-point rooted neighbor-joining dendrogram constructed with chord distances calculated from SNP data. All SNP data other than those from SIFA and SISP are from Clemento et al. (2014). Bootstrap values greater than 80% (out of 1000 bootstrap resamplings) are reported
Discussion
Here, we used genetic data to identify a previously unrecognized population of early returning (i.e., spring-run) Chinook salmon within the Siletz River, a basin that is <120 km from source to ocean exit. The use of three different types of molecular genetic data to investigate population genetic structure of Chinook salmon in this watershed allowed us to resolve fine-scale structure not previously recognized. Patterns of genetic variation within the watershed indicated that individuals spawning downstream of the waterfall (i.e., SILE and SIFA) are a fall-run population, and that a genetically distinct population of the spring-run ecotype (i.e., SISP) spawns upstream of the waterfall. Concordance among results from analyses of the multiple marker types lent strength to the resulting conclusion of significant structure in the Siletz River that corresponds to fish with different freshwater entry timing and to fish spawning above and below a waterfall. Pairwise FST values for populations spawning upstream and downstream of the waterfall were significant for all three molecular markers. The presumably neutral microsatellite loci that were used in this study were highly polymorphic and provided substantial power for resolution of genetic structure. However, persistent LD was found between some of these markers. The SNP markers were biallelic, and therefore less polymorphic per locus, but more numerous and did not have significant LD. The adaptive genetic markers correlate with behavioral variation in some salmon populations that have a temporal component and could therefore potentially discriminate such populations in the absence of other genetic differentiation. Previous work has shown that the combination of data from multiple marker types improved resolution of population structure in salmon, especially among populations with potentially high gene flow, as can be expected in smaller watersheds (Narum et al. 2008; Hess et al. 2011; DeFaveri et al. 2013; Garvin et al. 2013). Below, we discuss the patterns of genetic differentiation as inferred from the three sets of molecular markers and summarize the implications of our findings for continued conservation efforts.
Temporal variation
In contrast to the finding of significant differentiation between fall- and spring-run ecotypes returning above and below the waterfall with all three types of molecular marker, none of the datasets found differentiation between the lower river, early fall returning fish (SILE) and the lower river, fall-run ecotype (SIFA). There is no physical barrier between these two groups, but the timing of their return to freshwater and spawning dates suggested that heritable behavioral differences might exist. None of the pairwise FST values between SIFA and SILE were significant, indicating that these two groups of fish are likely experiencing high levels of gene flow. However, significant exact tests for genic and genotypic frequencies may indicate slight differentiation associated with this early spawning phenotype at a locus (Ots 515) that has been found to be associated with spawn time (O'Malley et al. 2007). We did not identify fine-scale genic and genotypic diversity using Clock1b and Cry 3, but these markers may only be informative for adaptive variation across basins, as suggested by O'Malley and Banks (2008). Although the existence of an early fall-run is informally acknowledged in some coastal rivers, only two non-fall-run populations of Chinook salmon (two spring-run populations in the upper Umpqua River) are formally delineated in the Oregon Coastal Multi-Species Conservation and Management Plan for the purposes of management and recovery (ODFW 2014). The finding of a genetically unique, wild population of spring-run Chinook salmon in the Siletz River indicates that greater life history diversity exists within this species in these smaller coastal rivers.
The adaptive genetic markers provided signals of structure that were similar to the other types of genetic markers, although variation in them has been found in other studies to be strongly associated with temporal variation in migration by salmon (O'Malley et al. 2007). Clock genes are part of the molecular mechanism of long-term timekeeping for tracking season-specific activities in response to photoperiod in many animals (Leder et al. 2006; Bradshaw and Holzapfel 2007; Liedvogel et al. 2009). Timing of freshwater entry and reproductive maturity in salmonid fishes is a complex array of interrelated behavioral and physiological traits that has heritable components and exhibits phenotypic plasticity in response to environmental variability (Carlson and Seamons 2008; Abadia-Cardoso et al. 2013). Chinook salmon populations have been able to exploit a wide range of habitats because of evolution at this trait, often in the face of ongoing gene flow (Waples 2001; O'Malley et al. 2013). Using markers associated with the circadian clock gene network might have provided additional insight in the differentiation among these closely related groups and they did discriminate the spring-run population above the waterfall from all downstream fish, although with genetic differentiation similar to the other markers.
Conservation implications
It is important for species to maintain genetic variability in order to respond to dynamic environments. The extent and scale of intraspecific genetic diversity is therefore a crucial consideration from both conservation and management perspectives (Manel et al. 2010; Funk et al. 2012). Chinook salmon populations are characterized by hierarchical genetic structure (Seeb et al. 2007; Moran et al. 2013) with evidence of isolation across their range in the north Pacific (> 10,000 km), within larger river systems (> 1000 km), and regionally among watersheds (< 1000 km). Finer-scale structure in Chinook salmon within smaller coastal watersheds (e.g., the Siletz River <120 km) has been relatively unstudied.
Matala et al. (2012) identified spatial structuring of Chinook salmon in the South Fork Salmon River (SFSR, ~90 km), which is part of the Columbia River system. Chinook salmon that spawned within the main stem SFSR were significantly different from individuals returning to two other tributaries within the subbasin. To reach these isolated tributaries, salmon must travel hundreds of kilometers up the main stem Columbia River. The FST values that we report between fish above and below the waterfall in the Siletz River are of similar magnitude to those reported among these Columbia River tributaries, but on a much smaller geographic scale. This demonstrates that such life history variation and genetic differentiation is not limited to large river systems and can be found in smaller watersheds.
Population structure described solely on the basis of divergence at one type of molecular marker, particularly presumably neutral ones, may fail to identify distinct populations that warrant separate management. Life history diversity of salmon is most often associated with spatial diversity and larger river systems (e.g., Columbia River or Sacramento-San Joaquin river systems) that typically have both numerous genetically distinct salmon populations and a greater number of life history strategies associated with them (Groot and Margolis 1991; Taylor 1991; Waples 2001).
Identifying and preserving genetic and phenotypic diversity is an important component of formulating strategies to maintain resiliency and fitness of salmon, particularly in smaller watersheds (McElhany et al. 2000). It has become evident that genetic data can inform many decisions relating to management strategies, especially those that are aimed at maintaining abundance and genetic diversity in natural salmon populations (Olsen et al. 2000; Eldridge and Killebrew 2007; Grandjean et al. 2009; Brenkman et al. 2012; Matala et al. 2012). Without such information there is great risk of losing important life history variation that enables resilience of anadromous species to changing environments.
Conclusions
We have demonstrated how the application of molecular genetic data from multiple types of markers provides strong support for existence of two genetically and phenotypically distinct salmon populations in a small coastal watershed where only one is currently recognized. Much of fishery management and conservation is based upon status of larger, regional management units (e.g., the North Oregon Coast Chinook Salmon ESU). These management units are combinations of unique spawning groups from multiple, smaller river basins (e.g., the Siletz, Alsea, Coquille, and Siuslaw Rivers). Management solely at such larger scales may not take into account fine-scale genetic and phenotypic variability that is present within smaller watersheds, such as has been demonstrated here. This fine-scale variability is a necessary component of long-term resilience and its maintenance should be explicitly considered to ensure successful conservation and management.
References
Abadia-Cardoso A, Anderson EC, Pearse DE, Garza JC (2013) Large-scale parentage analysis reveals reproductive patterns and heritability of spawn timing in a hatchery population of steelhead (Oncorhynchus mykiss). Mol Ecol 22:4733–4746. doi:10.1111/mec.12426
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (1996-2004) Genetix 4.05, 4.05 edn. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier
Bradshaw WE, Holzapfel CM (2007) Evolution of animal photoperiodism. Annu Rev Ecol Evol Syst 38:1–25. doi:10.1146/annurev.ecolsys.37.091305.110115
Brenkman SJ, Duda JJ, Torgersen CE, Welty E, Pess GR, Peters R, McHenry ML (2012) A riverscape perspective of Pacific salmonids and aquatic habitats prior to large-scale dam removal in the Elwha River, Washington, USA. Fish Manag Ecol 19:36–53. doi:10.1111/j.1365-2400.2011.00815.x
Carlson SM, Seamons TR (2008) A review of quantitative genetic components of fitness in salmonids: implications for adaptation to future change. Evol Appl 1:222–238. doi:10.1111/j.1752-4571.2008.00025.x
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Hum Genet 19:233–257
Clemento AJ, Crandall ED, Garza JC, Anderson EC (2014) Evaluation of a single nucleotide polymorphism baseline for genetic stock identification of Chinook Salmon (Oncorhynchus tshawytscha) in the California current large marine ecosystem. Fish Bull 112:112–130. doi:10.7755/fb.112.2-3.2
DeFaveri J, Viitaniemi H, Leder E, Merila J (2013) Characterizing genic and nongenic molecular markers: comparison of microsatellites and SNPs. Mol Ecol Resour 13:377–392. doi:10.1111/1755-0998.12071
Eldridge WH, Killebrew K (2007) Genetic diversity over multiple generations of supplementation: an example from Chinook salmon using microsatellite and demographic data. Conserv Genet 9:13–28. doi:10.1007/s10592-007-9298-y
Felsenstein J (1993) Phylip (phylogeny inference package), version 3.69 edn. Depatment of genetics, University of Washington, Seattle
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW (2012) Harnessing genomics for delineating conservation units. Trends Ecol Evol 27:489–496. doi:10.1016/j.tree.2012.05.012
Garvin MR et al (2013) Recent physical connections may explain weak genetic structure in western Alaskan chum salmon (Oncorhynchus keta) populations. Ecology and Evolution 3:2362–2377. doi:10.1002/ece3.628
Grandjean F, Verne S, Cherbonnel C, Richard A (2009) Fine-scale genetic structure of Atlantic salmon (Salmo salar) using microsatellite markers: effects of restocking and natural recolonization. Freshw Biol 54:417–433. doi:10.1111/j.1365-2427.2008.02116.x
Groot C, Margolis L (1991) Pacific Salmon life histories. University of British Columbia press, University of British Columbia, Vancouver, British Columbia
Hedrick PW (1999) Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53:313–318
Hess JE, Matala AP, Narum SR (2011) Comparison of SNPs and microsatellites for fine-scale spplication of genetic stock identification of Chinook salmon in the Columbia river basin. Mol Ecol Resour 11:137–149. doi:10.1111/j.1755-0998.2010.02958.x10.5061/dryad.7986
Ivanova NV, deWaard JR, Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6:1–5. doi:10.1111/j.1471-8286.2006.01428.x
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. doi:10.1111/j.1471-8286.2004.00845.x
Kinziger AP, Hellmair M, Hankin DG, Garza JC (2013) Contemporary population structure in Klamath River basin Chinook salmon revealed by analysis of microsatellite genetic data. Trans Am Fish Soc 142:1347–1357. doi:10.1080/00028487.2013.806351
Leder EH, Danzmann RG, Ferguson MM (2006) The candidate gene, clock, localizes to a strong spawning time quantitative trait locus region in rainbow trout. J Hered 97:74–80. doi:10.1093/jhered/esj004
Liedvogel M, Szulkin M, Knowles SC, Wood MJ, Sheldon BC (2009) Phenotypic correlates of clock gene variation in a wild blue tit population: evidence for a role in seasonal timing of reproduction. Mol Ecol 18:2444–2456. doi:10.1111/j.1365-294X.2009.04204.x
Lindley ST et al. (2004) Population structure of threatened and endangered Chinook salmon ESUs in California's Central Valley basin NOAA-NMFS-SWFSC-360. National Marine Fisheries Service, Southwest Fisheries Science Center
Manel S et al (2010) Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol Ecol 19:3760–3772. doi:10.1111/j.1365-294X.2010.04717.x
Matala AP, Narum SR, Young W, Vogel JL (2012) Influences of hatchery supplementation, spawner distribution, and habitat on genetic structure of Chinook salmon in the South fork Salmon River, Idaho. N Am J Fish Manag 32:346–359. doi:10.1080/02755947.2012.678961
McElhany P, Ruckelshaus MH, Ford MJ, Wainwright TC, Bjorkstedt EP (2000) Viable salmonid populations and the recovery of evolutionarily significant units. NOAA-NMFS-NWFSC-42.
Milner GB, Teel DJ, Utter F, Winans GA (1985) A genetic method of stock identification in mixed populations of Pacific salmon, Oncorhynchus spp. Mar Fish Rev 47:1–8
Moran P et al (2013) Divergent life-history races do not represent Chinook salmon coast-wide: the importance of scale in quaternary biogeography. Can J Fish Aquat Sci 70:415–435. doi:10.1139/cjfas-2012-0135
Naish KA, Park LK (2002) Linkage relationships for 35 new microsatellite loci in Chinook salmon Oncorhynchus tshawytscha. Anim Genet 33:312–318. doi:10.1046/j.1365-2052.2002.t01-4-00886.x
Narum SR et al (2008) Differentiating salmon populations at broad and fine geographical scales with microsatellites and single nucleotide polymorphisms. Mol Ecol 17:3464–3477. doi:10.1111/j.1365-294X.2008.03851.x
Nelson RJ, Beacham TD (1998) Isolation and cross species amplification of microsatellite loci useful for study of Pacific salmon. Anim Genet 30:225–244. doi:10.1046/j.1365-2052.1999.00404-4.x
Olsen JB, Bentzen P, Banks MA, Shaklee JB, Young S (2000) Microsatellites reveal population identity of individual pink salmon to allow supportive breeding of a population at risk of extinction. Trans Am Fish Soc 129:232–242. doi:10.1577/1548-8659(2000)129<0232:mrpioi>2.0.co;2
O'Malley KG, Banks MA (2008) A latitudinal cline in the Chinook salmon (Oncorhynchus tshawytscha) clock gene: evidence for selection on PolyQ length variants. Proceedings of the Royal Society B-Biological Sciences 275:2813–2821. doi:10.1098/rspb.2008.0524
O'Malley KG, Camara MD, Banks MA (2007) Candidate loci reveal genetic differentiation between temporally divergent migratory runs of Chinook salmon (Oncorhynchus tshawytscha). Mol Ecol 16:4930–4941. doi:10.1111/j.1365-294X.2007.03565.x
O'Malley KG, McClelland EK, Naish KA (2010) Clock genes localize to quantitative trait loci for stage-specific growth in juvenile coho salmon, Oncorhynchus kisutch. J Hered 101:628–632. doi:10.1093/jhered/esq063
O'Malley KG, Jacobson DP, Kurth R, Dill AJ, Banks MA (2013) Adaptive genetic markers discriminate migratory runs of Chinook salmon (Oncorhynchus tshawytscha) amid continued gene flow. Evol Appl 6:1184–1194. doi:10.1111/eva.12095
Oregon Department of Fish and Wildlife (1997) Siletz river basin fish management plan. Oregon Department of fish and Wildlife. Salem, Oregon
Oregon Department of Fish and Wildlife (2014) Coastal multi-species conservation and management plan. Oregon Department of Fish and Wildlife. Salem, Oregon
Peakall R, Smouse P (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research - an update. Bioinformatics:1-3 doi:10.1093/bioinformatics/bts460
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quinn TP (2004) The behavior and ecology of Pacific Salmon and Trout. University of Washington Press, Seattle
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Seeb LW et al (2007) Develoment of a standardized DNA database for Chinook salmon. Fisheries 31:540–552
Shafer ABA et al (2015) Genomics and the challenging translation into conservation practice. Trends Ecol Evol 30:78–87. doi:10.1016/j.tree.2014.11.009
Taylor EB (1991) A review of local adaptation in Salmonidae, with particular reference to Pacific and Atlantic salmon. Aquaculture 98:185–207
Utter FM, Hodgins HO (1972) Biochemical genetic variation at six loci in four stocks of rainbow trout. Trans Am Fish Soc 101:494–502. doi:10.1577/1548-8659(1972)101<494:bgvasl>2.0.co;2
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Waples RS (1998) Evolutionarily significant units, distinct population segments, and the endangered species act: reply to Pennock and Dimmick. Conserv Biol 12:718–721. doi:10.1111/j.1523-1739.1998.97524.x
Waples R (2001) Characterizing diversity in salmon from the Pacific northwest. J Fish Biol 59:1–41. doi:10.1006/jfbi.2001.1764
Waples RS, Teel DJ, Myers JM, Marshall AR (2004) Life-history divergence in Chinook salmon: historic contingency and parallel evolution. Evolution 58:386–403. doi:10.1554/03-323
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi:10.2307/2408641
Williamson KS, Cordes JF, May B (2002) Characterization of microsatellite loci in Chinook salmon (Oncorhynchus tshawytscha) and cross-species amplification in other salmonids. Mol Ecol Notes 2:17–19. doi:10.1046/j.1471-8286.2002.00129.x
Yoshiyama RM, Fisher FW, Moyle PB (1998) Historical abundance and decline of Chinook salmon in the Central Valley region of California. N Am J Fish Manag 18:487–521. doi:10.1577/1548-8675(1998)018<0487:HAADOC>2.0.CO;2
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 150 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Davis, C.D., Garza, J.C. & Banks, M.A. Identification of multiple genetically distinct populations of Chinook salmon (Oncorhynchus tshawytscha) in a small coastal watershed. Environ Biol Fish 100, 923–933 (2017). https://doi.org/10.1007/s10641-017-0616-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10641-017-0616-z