Abstract
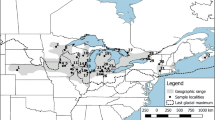
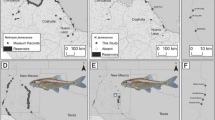
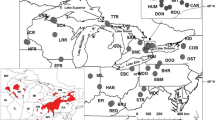
Blanding’s turtle, Emys blandingii, is a globally endangered species with a range centred on the Great Lakes of North America. Several disjunct populations also occur along the East Coast of North America. Previous studies suggest that the Great Lakes portion of the species’ range exhibits panmixia. However, E. blandingii is restricted to relatively small populations in many areas around the Great Lakes. Therefore, panmixia across large geographic distances in this area is unlikely. Here, we apply Bayesian analyses of population structure to samples collected across southern Ontario (N = 97) to test a null hypothesis of panmixia and assess possible management units (MUs), and to estimate rates of gene flow across the study area. Sampled sites in Ontario represent a minimum of four distinct genetic clusters of E. blandingii, which we recommend should be considered as independent MUs. Preliminary evidence suggests that further structure may be present in less robustly sampled areas, which deserve further consideration. Genetic diversity at sampled sites is comparable to that reported for other freshwater turtles. Our comparison between this study and previous work confirms that genetic diversity in E. blandingii is reduced in disjunct eastern populations compared to populations centred on the Great Lakes. Genetic diversity in E. blandingii is not correlated with latitude, and instead may reflect post-glacial dispersal of this species from multiple Pleistocene glacial refugia.



Similar content being viewed by others
References
Banning-Anthonysamy WJ (2012) Spatial ecology, habitat use, genetic diversity, and reproductive success: measures of connectivity of a sympatric freshwater turtle assemblage in a fragmented landscape. Dissertation, University of Illinois at Urbana-Champaign
Barton NH, Slatkin M (1986) A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56:409–416
Bleakney JS (1958) A zoogeographic study of the amphibians and reptiles of eastern Canada. Natl Mus Can Bull 155:1–119
Brecke BJ, Moriarty JJ (1989) Emydoidea blandingii (Blanding’s turtle). Longevity. Herpetol Rev 20:53
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Chiucchi JE, Gibbs HL (2010) Similarity of contemporary and historical gene flow among highly fragmented populations of an endangered rattlesnake. Mol Ecol 19:5345–5358
Congdon JD, van Loben Sels RC (1991) Growth and body size in the Blanding’s turtles (Emydoidea blandingii): relationships to reproduction. Can J Zool 69:239–245
Congdon JD, Dunham AE, van Loben Sels RC (1993) Delayed sexual maturity and demographics of Blanding’s turtles (Emydoidea blandingii): implications for conservation and management of long-lived organisms. Conserv Biol 7:826–833
Congdon JD, Graham TE, Herman TB, Lang JW, Pappas MJ, Brecke BJ (2008) Emydoidea blandingii (Holbrook 1838)—Blanding’s turtle. In: Rhodin AGJ, Pritchard PCH, van Dijk PP, Saumure RA, Buhlmann KA, Iverson JB (eds) Conservation biology of freshwater turtles and tortoises: a compilation project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelon Res Monogr 5:015.1–015.12. doi:10.3854/crm.5.015.blandingii.v1.2008. http://www.iucn-tftsg.org/cbftt/. Accessed 4 Feb 2013
COSEWIC (2005) COSEWIC assessment and update status report on the Blanding’s Turtle Emydoidea blandingii in Canada. Committee on the Status of Endangered Wildlife in Canadal, Ottawa, viii +40 pp. www.sararegistry.gc.ca/status/status_e.cfm. Accessed 10 Jan 2012
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Res 10:556–557
Davy CM, Leifso AE, Conflitti IM, Murphy RW (2012) Characterization of 10 novel microsatellite loci and cross-amplification of two loci in the snapping turtle (Chelydra serpentina). Conserv Genet Resour 4:695–698
Durand E, Jay F, Gaggiotti OE, François O (2009) Spatial inference of admixture proportions and secondary contact zones. Mol Biol Evol 26:1963–1973
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ernst CH, Lovich JL (2009) Turtles of the United States and Canada, 2nd edn. Johns Hopkins University, Baltimore
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stevens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1615
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University, Cambridge
Galbraith DA (2008) Population biology and population genetics. In: Steyermark AC, Finkler MS, Brooks RJ (eds) The biology of the snapping turtle. Johns Hopkins University, Baltimore, pp 168–180
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Green DM (2005) Designatable units for status assessment of endangered species. Conserv Biol 19:1813–1820
Holman JA (1992) Late quaternary herpetofauna of the central Great Lakes region, U.S.A.: zoogeographical and paleoecological implications. Quat Sci Rev 11:345–351
Holbrook JE (1838) North American herpetology; or, a description of the reptiles inhabiting the United States. In: Dobson J (ed), vol 3, 1st edn. Philadelphia, pp 122
Howes BJ, Brown JW, Gibbs HL, Herman TB, Mockford SW, Prior KA, Weatherhead PJ (2009) Directional gene flow patterns in disjunct populations of the black ratsnake (Pantherophis obsoletus) and the Blanding’s turtle (Emydoidea blandingii). Conserv Genet 10:407–417
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13. v.3.23. http://ibdws.sdsu.edu/. Accessed 10 Jan 2011
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Jost L (2009) D vs. GST: response to Heller and Siegismund (2009) and Ryman and Leimar. Mol Ecol 18:2088–2091
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kalinowski ST (2005) HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
King TL, Julian SE (2004) Conservation of microsatellite DNA flanking sequences across 13 Emydid genera assayed with novel bog turtle (Glyptemys muhlenbergii) loci. Conserv Genet 5:719–725
Mockford SW, Snyder M, Herman TB (1999) A preliminary examination of genetic variation in a peripheral population of Blanding’s turtle, Emydoidea blandingii. Mol Ecol 8:323–327
Mockford SW, McEachern L, Herman TB, Snyder M, Wright JM (2005) Population genetic structure of a disjunct population of Blanding’s turtle (Emydoidea blandingii) in Nova Scotia, Canada. Biol Conserv 123:373–380
Mockford SW, Herman TB, Snyder M, Wright JM (2007) Conservation genetics of Blanding’s turtle and its application in the identification of evolutionarily significant units. Conserv Genet 8:209–219
Moritz C (1994) Defining ‘evolutionarily significant units’ for conservation. Trends Ecol Evol 9:373–375
Osentoski MF, Mockford S, Wright JM, Snyder M, Herman TB, Hughes CR (2002) Isolation and characterization of microsatellite loci from the Blanding’s turtle, Emydoidea blandingii. Mol Ecol Notes 2:147–149
Paetkau D, Slade R, Burden M, Estoup A (2004) Direct, real-time estimation of migration rate using assignment methods: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Palsbøll PJ, Bérubé M, Allendorf FW (2006) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol 6:288–295
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GeneClass2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Power TD (1989) Seasonal movements and nesting ecology of a relict population of Blanding’s turtles (Emydoidea blandingii) in Nova Scotia. M.Sc. Thesis, Acadia University, Wolfville
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9221
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rosen T (2007) The endangered species act and the distinct population segment policy. Ursus 18:109–116
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Rubin CS, Warner RE, Bouzat JL, Paige KN (2001) Population genetic structure of Blanding’s turtles (Emydoidea blandingii) in an urban landscape. Biol Conserv 99:323–330
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning—a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Smith LM, Burgoyne LA (2004) Collecting, archiving and processing DNA from wildlife samples using FTA® databasing paper. BMC Ecol 4:4. http://www.biomedcentral.com/1472-6785/4/4. Accessed 12 Nov 2010
Templeton AR (1986) Coadaptation and outbreeding depression. In: Soulé M (ed) Conservation biology: the science of scarcity and diversity. Sinauer, Sunderland, pp 105–116
van Dijk PP, Rhodin AGJ (2011) Emydoidea blandingii. In: IUCN 2011. IUCN red list of threatened species. Version 2011.2. http://www.iucnredlist.org. Accessed 15 Dec 2011
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vargas-Ramirez M, Stuckas H, Castaňo-Mora OV, Fritz U (2012) Extremely low genetic diversity and weak population differentiation in the endangered Colombian river turtle Podocnemis lewyana (Testudines: podocnemididae). Conserv Genet 13:65–77
Waples RS (1991) Pacific salmon, Oncorhynchus spp. and the definition of “species” under the endangered species act. Mar Fish Rev 53:11–22
Whitlock MC, McCaughley DE (1999) Indirect measures of gene flow and migration: FST ≠ 1/(4 Nm + 1). Heredity 82:117–125
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
Laboratory analyses were funded by a Species at Risk Research Fund for Ontario grant from the Government of Ontario, and we thank B. Johnson and J. Philips of the Toronto Zoo for collaborating on this grant. Other funding was provided by a Canada Graduate Scholarship from the National Science and Engineering Research Council of Canada (NSERC) to CMD, a NSERC Canada Discovery Grant (A3148) to RWM and a Canada Collection grant from Wildlife Preservation Canada to CMD. Sample collection was possible thanks to the generous assistance of J. Baxter-Gilbert, S. Carstairs, B. Caverhill, S. Coombes, J. Crowley, J. Litzgus, M. Keevil, I. Macintosh, J. Paterson, J. Trottier, J. Riley, J. Rouse, D. Seburn, J. Urquhart and A. Whitear. J. Hathaway and J. Pierce allowed sampling of E. blandingii at Scales Nature Park. Jessica Hsiung assisted with figure preparation and provided an original illustration of a Blanding’s Turtle. We thank D. Currie, M. J. Fortin, D. McLennan, S. Lougheed, C. Wilson and two anonymous reviewers for comments on an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Davy, C.M., Bernardo, P.H. & Murphy, R.W. A Bayesian approach to conservation genetics of Blanding’s turtle (Emys blandingii) in Ontario, Canada. Conserv Genet 15, 319–330 (2014). https://doi.org/10.1007/s10592-013-0540-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0540-5