Abstract
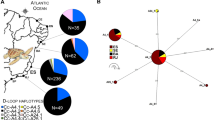
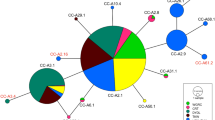
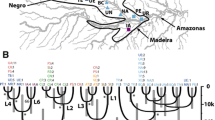
This study presents a comprehensive genetic analysis of stock structure for leatherback turtles (Dermochelys coriacea), combining 17 microsatellite loci and 763 bp of the mtDNA control region. Recently discovered eastern Atlantic nesting populations of this critically endangered species were absent in a previous survey that found little ocean-wide mtDNA variation. We added rookeries in West Africa and Brazil and generated longer sequences for previously analyzed samples. A total of 1,417 individuals were sampled from nine nesting sites in the Atlantic and SW Indian Ocean. We detected additional mtDNA variation with the longer sequences, identifying ten polymorphic sites that resolved a total of ten haplotypes, including three new variants of haplotypes previously described by shorter sequences. Population differentiation was substantial between all but two adjacent rookery pairs, and F ST values ranged from 0.034 to 0.676 and 0.004 to 0.205 for mtDNA and microsatellite data respectively, suggesting that male-mediated gene flow is not as widespread as previously assumed. We detected weak (F ST = 0.008 and 0.006) but significant differentiation with microsatellites between the two population pairs that were indistinguishable with mtDNA data. POWSIM analysis showed that our mtDNA marker had very low statistical power to detect weak structure (F ST < 0.005), while our microsatellite marker array had high power. We conclude that the weak differentiation detected with microsatellites reflects a fine scale level of demographic independence that warrants recognition, and that all nine of the nesting colonies should be considered as demographically independent populations for conservation. Our findings illustrate the importance of evaluating the power of specific genetic markers to detect structure in order to correctly identify the appropriate population units to conserve.



Similar content being viewed by others
References
Abreu-Grobois FA, Horrocks JA, Formia A, Dutton PH, LeRoux RA, Velez-Zuazo X, Soares L, Meylan AB (2006) New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis. In: Frick M, Panagopoulous A, Rees AF Williams K (eds) Proceedings of the 26th annual symposium on sea turtle biology. NOAA, Myrtle Beach, p 179
Augustinus PGE (2004) The influence of the trade winds on the coastal development of the Guianas at various scale levels: a synthesis. Mar Geol 208:145–151
Avise JC (1998) Conservation genetics in the marine realm. J Hered 89:377–382
Bandelt H, Forster P (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Billes A, Fretey J, Verhage B, Huijbregts B, Giffoni B, Prosdocimi L, Albareda DA, Georges J-Y, Tiwari M (2006) First evidence of leatherback movement from Africa to South America. Mar Turtle News 111:13–14
Bowen BW, Bass AL, Soares L, Toonen RJ (2005) Conservation implications of complex population structure: lessons from the loggerhead turtle (Caretta caretta). Mol Ecol 14:2389–2402
Carreras C, Pascual M, Cardona L, Aguilar A, Margaritoulis D, Rees AF, Turkozan O, Levy Y, Gasith A, Aureggi M, Khalil M (2007) The genetic structure of the loggerhead sea turtle (Caretta caretta) in the Mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conserv Genet 8:761–775
Chacón-Chaverri D, Eckert KL (2007) Leatherback sea turtle nesting at Gandoca Beach in Caribbean Costa Rica: management recommendations from fifteen years of conservation. Chelonian Conserv Biol 6:101–110
Chivers SJ, Dizon AE, Gearin PJ, Robertson KM (2002) Small-scale population structure of eastern North Pacific harbour porpoises (Phocoena phocoena) indicated by molecular genetic analyses. Fish Sci 4:111–122
DiBattista JD, Rocha LA, Craig MT, Feldheim KA, Bowen BW (2012) Phylogeography of two closely related Indo-Pacific butterflyfishes reveals divergent evolutionary histories and discordant results from mtDNA and microsatellites. J Hered 103(5):617–629
Dutton PH (1995) Molecular evolution of the sea turtles with special reference to the leatherback, Dermochelys coriacea. Ph.D. dissertation, Texas A&M University, College Station
Dutton PH (1996) Methods for collection and preservation of samples for sea turtle genetic studies. In: Bowen BW, Witzell WN (eds) Proceedings of the international symposium on sea turtle conservation genetics. NOAA technical memorandum NMFS-SEFSC-396. NOAA, Miami, p 17–24
Dutton PH, Frey A (2009) Characterization of polymorphic microsatellite markers for the green turtle (Chelonia mydas). Mol Ecol Resour 9:354–356
Dutton PH, Bowen BW, Owens DW, Barragan AR, Davis SK (1999) Global phylogeography of the leatherback turtle (Dermochelys coriacea). J Zool 248:397–409
Dutton DL, Dutton PH, Chaloupka M, Boulon RH (2005) Increase of a Caribbean leatherback turtle Dermochelys coriacea nesting population linked to long-term nest protection. Biol Conserv 126:186–194
Dutton PH, Hitipeuw C, Zein M, Benson SR, Al-Ghais SM (2007) Status and genetic structure of nesting populations of leatherback turtles (Dermochelys coriacea) in the Western Pacific. Chelonian Conserv Biol 6:47
Evans D, Ordonez C, Troeng S, Drews C (2007) Satellite tracking of leatherback turtles from Caribbean Central America reveals unexpected foraging grounds. In: Book of abstracts. Twenty-seventh annual symposium on sea turtle biology and conservation. NOAA, Miami, p 70–71
Excoffier L, Lischer HEL (2010) Arlequin suite ver. 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
FitzSimmons NN, Moritz C, Limpus CJ, Pope L, Prince RIT (1997) Geographic structure of the mitochondrial and nuclear gene polymorphisms in Australian green turtle populations and male-biased gene flow. Genetics 147:1843–1854
Girondot M, Fretey J (1996) Leatherback turtles, Dermochelys coriacea, nesting in French Guiana, 1978–1995. Chelonian Conserv Biol 2:204–208
Guo SW, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48:361–372
Hoelzel AR, Dover GA (1991) Genetic differentiation between sympatric Killer whale populations. Heredity 66:191–195
Hoffman JI, Matson CW, Amos W, Loughlin TR, Bickham JW (2006) Deep genetic subdivision within a continuously distributed and highly vagile marine mammal, the Steller’s sea lion (Eumetopias jubatus). Mol Ecol 15:2821–2832
IUCN (2009) IUCN red list of threatened species. Version 2009.2. www.iucnredlist.org. Accessed 1 March 2010
James MC, Eckert SA, Myers RA (2005) Migratory and reproductive movements of male leatherback turtles (Dermochelys coriacea). Mar Biol 147:845–853
Jensen MP, FitzSimmons NN, Dutton PH (in press) Molecular genetics of sea turtles. In: Musick J, Lohman K, Wyneken J (eds) The biology of sea turtles, vol 3. CRC Press, Boca Raton
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Karl SA, Bowen BW (1999) Evolutionary significant units versus geopolitical taxonomy: molecular systematics of an endangered sea turtle (Genus Chelonia). Conserv Biol 13:990–999
Karl SA, Bowen BW, Avise JC (1992) Global population genetic structure and male-mediated gene flow in the green turtle (Chelonia mydas): RFLP analyses of anonymous nuclear loci. Genetics 131:163–173
Karl SA, Toonen RJ, Grant WS, Bowen BW (2012) Common misconceptions in molecular ecology: echoes of the modern synthesis. Mol Ecol 21:4171–4189
LaCasella EL, Dutton PH (2008) Longer mtDNA sequences resolve leatherback stock structure. In: Rees AF, Frick M, Panagopoulou A, Williams K (eds.) Proceedings of the twenty-seventh annual symposium on sea turtle biology and conservation, p 128–129. NOAA technical memorandum NMFS-SEFSC-569. NOAA, Miami, p 262
Larsson LC, Charlier J, Laikre L, Ryman N (2009) Statistical power for detecting genetic divergence—organelle versus nuclear markers. Conserv Genet 10:1255–1264
LeRoux RA, Dutton PH, Abreu-Grobois FA, Lagueux CJ, Campbell CL, Delcroix E, Chevalier J, Horrocks JA, Hillis-Starr Z, Troëng S, Harrison E, Stapleton S (2012) Re-examination of population structure and phylogeography of hawksbill turtles in the Wider Caribbean using longer mtDNA sequences. J Hered 103(6):806–820
Liew HC (2011) Tragedy of the Malaysian leatherback population: what went wrong. In: Dutton PH, Squires D, Mahfuzuddin A (eds) Conservation and sustainable management of sea turtles in the Pacific Ocean. University of Hawaii Press, Hawaii, pp 97–106
Martinez LS, Barragan AR (2011) Importance of networks for conservation of the Pacific leatherback turtle: the case of “Projecto Laud” in Mexico. In: Dutton PH, Squires D, Mahfuzuddin A (eds) Conservation and sustainable management of sea turtles in the Pacific Ocean. University of Hawaii Press, Hawaii, pp 120–131
Meirmans PG, Hedrick PW (2011) Assessing population structure: Fst and related measures. Mol Ecol Resour 11:5–18
Mesnick S, Taylor B, Archer FI, Martien K et al (2011) Sperm whale population structure in the eastern North Pacific inferred by the use of single nucleotide polymorphisms (SNPs), microsatellites and mitochondrial DNA. Mol Ecol Resour 11:278–298
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res 16:1215
Monzón-Argüello C, Rico C, Naro-Maciel E, Varo-Cruz N, López P, Marco A, L-J LF (2010) Population structure and conservation implications for the loggerhead sea turtle of the Cape Verde Islands. Conserv Genet 11:1871–1884
Moritz C (1994) Defining “evolutionarily significant units” for conservation. Trends Ecol Evol 9:373–375
Mrosovsky N, Godley MH (2010) Thoughts on climate change and sex ratio of sea turtles. Mar Turtle News 128:7–11
O’Corry-Crowe GM, Suydam RS, Rosenberg A, Frost KJ, Dizon AE (1997) Phylogeography, population structure and dispersal patterns of the beluga whale Delphinapterus leucas in the western Nearctic revealed by mitochondrial DNA. Mol Ecol 6:955–970
O’Corry-Crowe GM, Taylor BL, Gelatt T, Loughlin TR, Bickham J, Basterretche M, Pitcher KW, DeMaster DP (2006) Demographic independence along ecosystem boundaries in Steller sea lions revealed by mtDNA analysis: implications for management of an endangered species. Can J Zool 84:1796–1809
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Roberts MA, Schwartz TS, Karl SA (2004) Global population genetic structure and male-mediated gene flow in the green sea turtle (Chelonia mydas): analysis of microsatellite loci. Genetics 166:1857
Roden SE, Dutton PH (2011) Isolation and characterization of 14 polymorphic microsatellite loci in the leatherback turtle (Dermochelys coriacea) and cross-species amplification. Conserv Genet Resour 3:49–52
Ryman N, Palm S (2006) POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes 6:600–602
Ryman N, Palm S, André C, Carvalho GR, Dahlgren TG, Jorde PE, Laikre L, Larsson LC, Palmé A, Ruzzante DE (2006) Power for detecting genetic divergence: differences between statistical methods and marker loci. Mol Ecol 15:2031–2045
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbour Laboratory Press, New York
Shamblin BM, Dodd MG, Bagley DA, Ehrhart LM, Tucker AD, Johnson C, Carthy RR, Scarpino RA, McMichael E, Addison DS et al (2011) Genetic structure of the southeastern United States loggerhead turtle nesting aggregation: evidence of additional structure within the peninsular Florida recovery unit. Mar Biol 158:1–17
Shamblin BM, Bolten AB, Bjorndal KA, Dutton PH, Nielsen JT, Abreu-Grobois FA, Reich KJ, Witherington BE, Bagley DA, Ehrhart LM, Tucker AD, Addison DS, Arenas A, Johnson C, Carthy RR, Lamont MM, Dodd MG, Gaines MS, LaCasella E, Nairn CJ (2012) Expanded mitochondrial control region sequences increase resolution of stock structure among North Atlantic loggerhead turtle rookeries. Mar Ecol Prog Ser 469:145–160
Stewart KR, Sims M, Meylan AB, Witherington BE, Brost B, Crowder LB (2011) Leatherback nests increasing significantly in Florida, USA; trends assessed over 30 years using multilevel modeling. Ecol Appl 21:263–273
Tapilatu RF, Dutton PH, Tiwari M, Wibbels T, Ferdinandus HV, Iwanggin WG, Nugroho BH (2013) Long-term decline of the western Pacific leatherback, Dermochelys coriacea; a globally important sea turtle population. Ecosphere (in press)
Taylor BL (1997) Defining ‘‘population’’ to meet management objectives for marine mammals. In: Dizon AE, Chivers SJ, Perrin WF (eds) Molecular genetics of marine mammals, special publication 3. Society for Marine Mammalogy, Lawrence, p 49–65
Taylor BL, Dizon AE (1999) First policy then science: why a management unit based solely on genetic criteria cannot work. Mol Ecol 8:S11–S16
Taylor BL, Martien K, Morin P (2010) Identifying units to conserve using genetic data. In: Boyd IL, Bowen WD, Iverson SJ (eds) Marine mammal ecology and conservation—a handbook of techniques. Oxford University Press, Oxford, p 306–344
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Troëng S, Chacón D, Dick B (2004) Possible decline in leatherback turtle Dermochelys coriacea nesting along the coast of Caribbean Central America. Oryx 38:395–403
Troëng S, Harrison E, Evans D, Haro AD, Vargas E (2007) Leatherback turtle nesting trends and threats at Tortuguero, Costa Rica. Chelonian Conserv Biol 6:117–122
Turtle Expert Working Group (2007) An assessment of the leatherback turtle population in the Atlantic Ocean. NOAA Technical Memorandum NMFS SEFSC-555 116. NOAA, Miami
Vargas SM, Araujo FC, Monteiro DS, Estima SC, Almeida AP, Soares LS, Santos FR (2008) Genetic diversity and origin of leatherback turtles (Dermochelys coriacea) from the Brazilian coast. J Hered 99:215
Velez-Zuazo X, Ramos WD, Van Dam RP, Diez CE, Abreu-Grobois FA, McMillan WO (2008) Dispersal, recruitment and migratory behaviour in a hawksbill sea turtle aggregation. Mol Ecol 17:839–853
Wallace BP, DiMatteo AD, Hurley BJ, Finkbeiner EM, Bolten AB, Chaloupka MY, Hutchinson BJ et al (2010) Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PLoS One 5:e15465
Wallace BP, DiMatteo AD, Bolten AB, Chaloupka MY, Hutchinson BJ et al (2011) Global conservation priorities for marine turtles. PLoS One 6(9):e24510. doi:10.1371/journal.pone.0024510
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC (2011) G′ST and D do not replace FST. Mol Ecol 20:1083–1091
Witt MJ, Baert B, Broderick AC, Formia A, Fretey J, Gibudi A, Mounguengui GAM, Moussounda C, Nougessono S, Parnell RJ, Roumet D, Sounguet G-P, Verhage S, Zogo A, Godley BJ (2009) Aerial surveying of the world’s largest leatherback turtle rookery: a more effective methodology for large-scale monitoring. Biol Conserv 142:1719–1727
Witt MJ, Bonguno EA, Broderick AC, Coyne MS, Formia A, Gibudi A, Mounguengui GAM, Moussounda C, Nsafou M, Nougessono S, Parnell RJ, Sounguet G-P, Verhage S, Godley BJ (2011) Tracking leatherback turtles from the world’s largest rookery: assessing threats across the South Atlantic. Proc R Soc B 278:2338–2347
Acknowledgments
Samples used in this study are archived in the US National Marine Fisheries Service (NMFS) Marine Turtle Molecular Research Sample Collection at the Southwest Fisheries Science Center and were collected under the respective national authorizations and CITES permit conditions and imported under CITES permit 844694/9. We thank all of the members of the SWFSC Marine Turtle Research Program for their support, including Robin LeRoux, Amy Frey, Vicki Pease, Amy Lanci, Gabriela Serra-Valente and Amanda Bowman, who assisted with laboratory analysis. Special thanks to Eric Archer for help with statistical analysis. For providing samples we thank US Fish and Wildlife Service, Sandy Point National Wildlife Refuge, US Virgin Islands Department of Planning and Natural Resources, Natal Parks Board, Costa Rica Conservation Areas System (SINAC), ANAI, CONAGEBIO, Nature Seekers, the PAWI Club, WIDECAST, Proyecto Tamar/ICMBio, Cecília Baptistotte, Antônio Almeida, Niki Desjardin, Chris Johnson, Marc Girondot, Jeremy Smith, Shana Phelan, Kelly Martin, Sandy Fournies, Jesse Marsh, George Hughes, Andrews Agyekumhene, Karyn Allman and Jeanne and Steve Garner. We are grateful to Bill Perrin, Michael Jensen, Brian Bowen and an anonymous reviewer for helpful comments on the manuscript. Research was funded by the NMFS. Fieldwork funded in part by Duke Marine Laboratory, Florida Turtle License Plate Fund, Loggerhead Marinelife Center, Andrea Simler, MacArthur Beach State Park, National Save the Sea Turtle Foundation, PADI Aware, Oak Foundation, the Darwin Initiative, University of Glasgow, Sigma Xi, Earthwatch, US Fish and Wildlife Service.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dutton, P.H., Roden, S.E., Stewart, K.R. et al. Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv Genet 14, 625–636 (2013). https://doi.org/10.1007/s10592-013-0456-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0456-0