Abstract
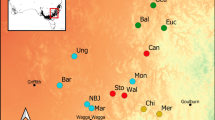
It is generally accepted that most plant populations are locally adapted. Yet, understanding how environmental forces give rise to adaptive genetic variation is a challenge in conservation genetics and crucial to the preservation of species under rapidly changing climatic conditions. Environmental variation, phylogeographic history, and population demographic processes all contribute to spatially structured genetic variation, however few current models attempt to separate these confounding effects. To illustrate the benefits of using a spatially-explicit model for identifying potentially adaptive loci, we compared outlier locus detection methods with a recently-developed landscape genetic approach. We analyzed 157 loci from samples of the alpine herb Gentiana nivalis collected across the European Alps. Principle coordinates of neighbor matrices (PCNM), eigenvectors that quantify multi-scale spatial variation present in a data set, were incorporated into a landscape genetic approach relating AFLP frequencies with 23 environmental variables. Four major findings emerged. 1) Fifteen loci were significantly correlated with at least one predictor variable (R 2adj > 0.5). 2) Models including PCNM variables identified eight more potentially adaptive loci than models run without spatial variables. 3) When compared to outlier detection methods, the landscape genetic approach detected four of the same loci plus 11 additional loci. 4) Temperature, precipitation, and solar radiation were the three major environmental factors driving potentially adaptive genetic variation in G. nivalis. Techniques presented in this paper offer an efficient method for identifying potentially adaptive genetic variation and associated environmental forces of selection, providing an important step forward for the conservation of non-model species under global change.




Similar content being viewed by others
References
Aeschimann D, Lauber K, Moser DM, Theurillat J (2004) Flora alpina, vol 2. Haupt, Berne
Allendorf FW, Hohenlohe PA, Luikart G (2010) Genomics and the future of conservation genetics. Nat Rev Genet 11:697–709
Alvarez N, Manel S, Schmitt T, IntraBioDiv Consortium (2012) Contrasting diffusion of quaternary gene pools across Europe: the case of the arctic-alpine Gentiana nivalis L. (Gentianaceae). Flora 207:408–413
Alvarez N, Thiel-Egenter C, Tribsch A et al (2009) History or ecology? Substrate type as a major driver of spatial genetic structure in Alpine plants. Ecol Lett 12:632–640
Antao T, Beaumont MA (2011) MCHEZA: a workbench to detect selection using dominant markers. Bioinformatics 27:1717–1718
Barton NH (2000) Genetic Hitchhiking. Philos Trans R Soc Lond B 355:1553–1562
Beaumont MA, Balding DJ (2004) Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol 13:969–980
Beaumont MA, Nichols RA (1996) Evaluating loci for use in the genetic analysis of population structure. Proc Royal Soc London B 263:1619–1626
Bellier E, Monestiez P, Durbec J-P, Candau J-N (2007) Identifying spatial relationships at multiple scales: principal coordinates of neighbor matrices (PCNM) and geostatistical approaches. Ecography 30:385–399
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B 57:289–300
Bensch S, Åkesson M (2005) Ten years of AFLP in ecology and evolution: why so few animals? Mol Ecol 14:2899–2914
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2005) GenBank. Nucleic Acids Res. doi:10.1093/nar/gki063
Borcard D, Legendre P (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbor matrices. Ecol Model 153:51–68
Borcard D, Legendre P, Drapeau P (1992) Partialling out the spatial component of ecological variation. Ecology 73:1045–1055
Borcard D, Gillet F, Legendre P (2011) Numerical ecology with R. Springer, New York
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach. Springer, New York
Burnham KP, Anderson DR (2004) Multimodel inference: understanding AIC and BIC in model selection. Sociol Method Res 33:261–304
Coop G, Witonsky D, Di Rienzo A, Pritchard JK (2010) Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423
Crawford RMM (2008) Plants at the margin. Ecological limits and climate change. Cambridge University Press, Cambridge
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Endler JA (1977) Geographic variation, speciation and clines. Princeton University Press, Princeton
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Foll M, Gaggiotti O (2008) A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Fournier-Level A, Korte A, Cooper MD, Nordborg M, Schmitt J, Wilczek AM (2011) A map of local adaptation in Arabidopsis thaliana. Science 334:86–89
Gibson N, Yates CJ, Dillon R (2010) Plant communities of the ironstone ranges of South Western Australia: hotspots for plant diversity and mineral deposits. Biodiv Conserv 19:3951–3962
Gugerli Z, Englisch T, Niklfeld H et al (2008) Relationships among levels of biodiversity and the relevance of intraspecific diversity in conservation – a project synopsis. Perspect Plant Ecol Evol Syst 10:259–281
Hansen MM, Olivieri I, Waller DM, Nielsen EE (2012) The GeM Working Group. Monitoring adaptive genetic responses to environmental change. Mol Ecol. doi: 10.1111/j.1365-294X.2011.05463.x
Hegi G (1957) Illustrierte flora von Mittel-Europa, vol 3. Lehmanns, München
Hess HE, Landolt E, Hirzel R (1972) Flora der Schweiz und angrenzender Gebiete, vol 3. Birkhäuser, Basel
Hirao AS, Kudo G (2004) Landscape genetics of alpine-snowbed plants: comparisons along geographic and snowmelt gradients. Heredity 93:290–298
Hoffmann AA, Willi Y (2008) Detecting genetic responses to environmental change. Nat Rev Genet 9:421–432
Holderegger R, Wagner HH (2008) Landscape genetics. Bioscience 58:199–207
Holderegger R, Hermann D, Poncet B et al (2008) Land ahead: using genome scans to identify molecular markers of adaptive relevance. Plant Ecol Divers 1:273–283
Holderegger R, Bühler D, Gugerli F, Manel S (2010) Landscape genetics of plants. Trends Plant Sci 15:675–683
Hultén E, Fries M (1986) Atlas of North European vascular plants: north of the Tropic of Cancer. Koeltz, Königstein
Ingvarsson PK, García MV, Hall D, Luquez V, Jansson S (2006) Clinal variation in phyB2, a candidate gene for day-length-induced growth cessation and bud set, across a latitudinal gradient in European aspen (Populus tremula). Genetics 172:1845–1853
Jay F, Manel S, Alvarez N et al (2012) Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol Ecol 21:2354–2368
Jeffreys H (1961) The theory of probability, 3rd edn. Oxford University Press, New York, p 432
Jombart T, Dray S, Dufour A-B (2009) Finding essential scales of spatial variation in ecological data: a multivariate approach. Ecography 32:161–168
Joost S, Bonin A, Bruford MW et al (2007) A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol Ecol 16:3955–3969
Körner C (2003) Alpine plant life: functional plant ecology of high mountain ecosystems. Springer, New York
Kozuharova E, Anchev M (2002) Floral biology, pollination ecology and breeding systems in Gentiana verna, G. utriculosa and G. nivalis (sect. Calatianae, Gentianaceae). God Sofiisk Univ St. Kliment Ohridski Biol Fak 2 Bot 92:57–71
Kozuharova E, Anchev ME (2006) Nastic corolla movements of nine Gentiana species (Gentianaceae), presented in the Bulgarian flora. Phytol Balcanica 12:255–265
Legendre P, Legendre L (1998) Numerical ecology. Elsevier, Amsterdam
Manel S, Conord C. Despres L (2009) Genome scan to assess the respective role of host - plant and environmental constraints on the adaptation of a widespread insect. BMC. Evol Biol 9:288. http://www.biomedcentral.com/1471-2148/9/288
Manel S, Gugerli F, Thuiller W, Alvarez N, Legendre P, Holderegger R, Gielly L, Taberlet P, IntraBioDiv Consortium (2012) Broad-scale adaptive genetic variation in alpine plants is driven by temperature and precipitation. Mol Ecol 21:3729–3738
Manel S, Segelbacher G (2009) Perspectives and challenges in landscape genetics. Mol Ecol 18:1821–1822
Manel S, Berthier P, Luikart G (2002) Detecting wildlife poaching: identifying the origin of individuals with Bayesian assignment tests and multilocus genotypes. Conserv Biol 3:650–659
Manel S, Joost S, Epperson B et al (2010a) Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol Ecol 19:3760–3772
Manel S, Poncet NB, Legendre P, Gugerli F, Holderegger R (2010b) Common factors drive genetic variation of adaptive relevance at different spatial scales in Arabis alpina. Mol Ecol 19:3824–3835
Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses and advances. Trends Plant Sci 12:106–117
Nielsen R (2005) Molecular signatures of natural selection. Annu Rev Genet 39:197–218
Ohtani K (2000) Bootstrapping R2 and adjusted R2 in regression analysis. Econ Model 17:473–483
Okuda T, Noda T, Yamamoto T, Hori M, Nakaoka M (2010) Contribution of environmental and spatial processes to rocky intertidal metacommunity structure. Acta Oecol 36:413–422
Parisod C, Joost S (2010) Divergent selection in trailing- versus leading-edge populations of Biscutella laevigata. Ann Bot 105:655–660
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Poncet B, Herrmann D, Gugerli F et al (2010) Tracking genes of ecological relevance using a genome scan: application to Arabis alpina. Mol Ecol 19:2896–2907
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2008/2009) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. (http://www.R-project.org.)
Schmidt PS, Serrao EA, Pearson GA et al (2008) Ecological genetics in the north Atlantic: environmental gradients and adaptation at specific loci. Ecology 89:S91–S107
Schoville S, Bonin A, Francois O, Lobreaux S, Melodelima C, Manel S (2012) Adaptive genetic variation on the landscape: methods and cases. Annu Rev Ecol Evol Syst 43:23–43
Storz JF (2005) Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol 14:671–688
Taberlet P, Zimmermann NE, Englisch T et al (2012) Genetic diversity in widespread species is not congruent with species richness in alpine plant communities. Ecol Lett (in press)
Thuiller W (2007) Biodiversity: climate change and the ecologist. Nature 448:550–552
Wagner HH, Fortin M-J (2005) Spatial analysis of landscapes: concepts and statistics. Ecology 86:1975–1987
Walther GR, Post E, Convey P et al (2002) Ecological responses to recent climate change. Nature 416:389–395
Waples RS, Gaggiotti OE (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Acknowledgments
This research was made possible through the National Center for Ecological Analysis and Synthesis (NCEAS) at the University of California in Santa Barabara (Distributed Graduate Seminar on Landscape Genetics). We thank the IntraBioDiv Consortium for the use of their G. nivalis genetic data set. We are grateful to J. Bregy, D. Bühler, S. Dray, P. Legendre, the Cottonwood Ecology Group, and two anonymous reviewers for thoughtful discussions and comments on earlier versions of the manuscript. NA is funded by the Swiss National Science Foundation (Ambizione fellowship PZ00P3-126624). SM was supported by the Institut Universitaire de France.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bothwell, H., Bisbing, S., Therkildsen, N.O. et al. Identifying genetic signatures of selection in a non-model species, alpine gentian (Gentiana nivalis L.), using a landscape genetic approach. Conserv Genet 14, 467–481 (2013). https://doi.org/10.1007/s10592-012-0411-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0411-5