Abstract
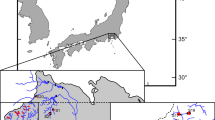
The amago salmon, Oncorhynchus masou ishikawae, is an endemic subspecies of O. masou in Japan. Populations in the River Miya, located at the entry of Ise Bay, central Japan, represent indigenous wild populations. They have been isolated in small tributaries as a result of environmental degradation in recent decades, with some populations also partially affected by stocking of hatchery fish of River Miya origin. We examined the genetic structure of these populations, using mitochondrial (mtDNA) and microsatellite DNA markers, in order to devise strategies for their long-term management and conservation. For mtDNA, sympatry of two genetically distinct clades was observed in some River Miya populations, together with populations from other rivers flowing into Ise Bay drainage. Taking into consideration paleogeographical information, it is conceivable that large-scale genetic admixture of the two clades in O. m. ishikawae occurred in Ise Bay in the past. For microsatellites, inter-population genetic divergence was small and genetic bottlenecks were detected in small-sized populations, with many microsatellite loci monomorphic. Although a Bayesian-based assignment test suggested two clusters in River Miya populations, they are likely to have been caused by anthropogenic effects in recent years since these clusters reflect population bottlenecks, not mtDNA clades. In conclusion, indigenous wild populations of O. m. ishikawae in the River Miya should be treated as a single conservation unit and the exchange of individuals between tributaries is needed to prevent the progress of genetic bottlenecks.




Similar content being viewed by others
References
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell Publishing, Malden
Allendorf FW, Waples RS (1996) Conservation and genetics of salmonid fishes. In: Avise JC, Hamrick JL (eds) Conservation genetics: case histories from nature. Chapman & Hall, New York, pp 238–280
Brooker AL, Benzie JAH, Blair D, Versini JJ (2000) Population structure of the giant tiger prawn Penaeus monodon in Australian waters, determined using microsatellite markers. Mar Biol 136:149–157
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Human Genet 19:233–257
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Cox CB, Moore PD (2005) Biogeography: an ecological and evolutionary approach. Blackwell Publishing, Malden
EL Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor App Gen 92:832–839
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Felsenstein J (2005) PHYLIP (Phylogeny inference package), Version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Goudet J (2001) FSTAT: a program to estimate and test gene diversities and fixation indices. Version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm. Accessed 30 Nov 2011
Kano Y, Tomida Y, Ikeda N, Iwawaki N, Miyagawa M, Harada Y, Ichiyanagi H, Watanabe K (2011) Fluctuation and variation in stream-fish assemblages after a catastrophic flood in the Miyagawa River, Japan. Environ Biol Fish 92:447–460
Kawamura K, Kubota M, Furukawa M, Harada Y (2007) The genetic structure of endangered indigenous populations of the amago salmon, Oncorhynchus masou ishikawae, in Japan. Conserv Genet 8:1163–1176
Kawamura K, Furukawa M, Kubota M, Harada Y (2012) Effects of stocking hatchery fish on the phenotype of indigenous populations in the amago salmon Oncorhynchus masou ishikawae in Japan. J Fish Biol. doi:10.1111/j.1095-8649.2012.03315.x
Kawanabe H (1989) Japanese char(r(r))s and masu-salmon problems: a review. Physiol Ecol Jpn Spec 1:13–24
Kinnison MT, Hendry AP (2004) From macro- to micro-evolution: tempo and mode in salmonid evolution. In: Hendry AP, Stearns SC (eds) Evolution illuminated: salmon and their relatives. Oxford University Press, New York, pp 208–231
Kudo H, Inoguchi N, Kijima A (2002) Estimation of the heritability of parr mark numbers by correlation between masou trout (Oncorhynchus masou) parents and offspring. Fish Genet Breed Sci 32:11–18
Lansman RA, Shade RO, Shapira JF, Avise JC (1981) The use of restriction endonucleases to measure mitochondrial DNA sequence relatedness in natural populations. III. Techniques and potential applications. J Mol Evol 17:214–226
Luikart G, Sherwin WB, Steele BM, Allendorf FW (1998) Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Mol Ecol 7:963–974
Martin A, Palumbi S (1993) Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci USA 90:4087–4091
Moritz C (1994) Defining ‘Evolutionarily Significant Units’ for conservation. Trends Ecol Evol 9:373–375
Moriyama A (2004) Submarine topography, especially the formation of caldrons and sand banks in the Ise and Mikawa Bay. Bull Aichi Univ Educ Natural Sci 53:39–56
Nakamura T, Iida H (2009) Conservation and enhancement of charr and salmon in Japanese mountain streams. Rural Culture Association Japan, Tokyo
Nakano S, Taguchi S, Shibata Y, Tanaka T (2001) Oncorhynchus masou ishikawae. In: Kawanabe H, Mizuno N, Hosoya K (eds) Freshwater fishes in Japan. Yama-kei, Tokyo, pp 169–178
Nawarathna B, Ishino K, Tachikawa Y, Tamai N (2006) Application of distributed hydrological modeling to identify river structures vulnerable to extreme floods. In. Proceedings of the 3rd APHW conference on “Wise Water Resources Management towards Sustainable Growth and Poverty Reduction”, Bangkok, p 8, 16–18 Oct 2006
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Noguchi D, Ikeda M, Nakajima M, Taniguchi N (2003) Isolation and characterization of microsatellite DNA markers for population genetics study of masu salmon, Oncorhnychus masou masou. Fish Genet Breed Sci 33:61–66
Olsen JB, Wenburg JK, Bentzen P (1996) Semiautomated multilocus genotyping of Pacific salmon (Oncorhnychus spp.) using microsatellites. Mol Mar Biol Biotech 5:259–272
Oshima M (1957) Studies on the dimorphic salmons, Oncorhynchus masou (Brevoort) and Oncorhynchus rhodurus Jordan and McGregor, found in Japan and adjacent territories. Nireshobou, Sapporo
Piry S, Luikart G, Cornuet J-M (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered 90:502–503
Poteaux C, Bonhomme F, Berrebi P (1999) Microsatellite polymorphism and genetic impact of restocking in Mediterranean brown trout (Salmo trutta L.). Heredity 82:645–653
Raymond M, Rousset F (1995) GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Ryder OA (1986) Species conservation and systematics: the dilemma of subspecies. Trends Ecol Evol 1:9–10
Scribner KT, Gust JR, Fields RL (1996) Isolation and characterization of novel salmon microsatellite loci: cross-species amplification and population genetic applications. Can J Fish Aquat Sci 53:833–841
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Tachikawa W, Kumazaki T (1981) Studies on the reproduction of amago salmon, Oncorhynchus rhodurus-XXI. On the hereditary tendency of the external red spots of amago salmon, Oncorhynchus rhodurus, with reference to those number and size. Rep Gifu Prefec Fish Exp Stn 26:1–4
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waples RS, McClure MM, Wainwright TC, McElhany P, Lawson PW (2010) Integrating evolutionary considerations into recovery planning for Pacific salmon. In: DeWoody JA, Bickham JW, Michler C (eds) Molecular approaches in natural resource conservation and management. Cambridge University Press, New York, pp 238–280
Watanabe K, Mori S (2008) Comparison of genetic population structure between two cyprinids, Hemigrammocypris rasborella and Pseudorasbora pumila subsp., in the Ise Bay basin, central Honshu, Japan. Ichthyol Res 55:309–320
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Yamamoto S, Morita K, Koizumi I, Maekawa K (2004) Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: spatial-temporal changes in gene frequencies. Conserv Genet 5:529–538
Yonezawa J, Hasegawa A, Saitoh S, Sato R (2000) Heredity of the number of parr marks and black spots in masu salmon, Oncorhynchus masou masou. Rep Tokyo Metropol Fish Exp Stn 212:7–11
Acknowledgments
We would like to thank Y. Kano, I. Murakami, T. Tokuhara, and the stuff of the Laboratory of Marine Population Dynamics, Faculty of Bioresources, Mie University for their help in collecting samples. We also want to express our sincere thanks to Y. Mizutani, K. Kawabata, Y. Nishi, and Miyagawa-jyoryu Fisheries Cooperative for their efficient assistance and provision of vital information. We are also grateful to C. Smith for comments and suggestion. This work was carried out as one of the projects for the stock enhancement of landlocked salmonid fishes, organized by Fisheries Agency in Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miyahara, H., Yamada, H., Sato, T. et al. Mitochondrial–nuclear discordance in the amago salmon, Oncorhynchus masou ishikawae, in the River Miya, Japan. Conserv Genet 13, 1343–1353 (2012). https://doi.org/10.1007/s10592-012-0378-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0378-2