Abstract
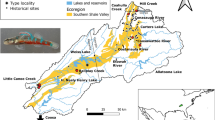
Imperiled Okaloosa darters (Etheostoma okaloosae) are small, benthic fish limited to six streams that flow into three bayous of Choctawhatchee Bay in northwest Florida, USA. We analyzed the complete mitochondrial cytochrome b gene and 10 nuclear microsatellite loci for 255 and 273 Okaloosa darters, respectively. Bayesian clustering analyses and AMOVA reflect congruent population genetic structure in both mitochondrial and microsatellite DNA. This structure reveals historical isolation of Okaloosa darter streams nested within bayous. Most of the six streams appear to have exchanged migrants though they remain genetically distinct. The U.S. Fish and Wildlife Service recently reclassified Okaloosa darters from endangered to threatened status. Our genetic data support the reclassification of Okaloosa darter Evolutionary Significant Units (ESUs) in the larger Tom’s, Turkey, and Rocky creeks from endangered to threatened status. However, the three smaller drainages (Mill, Swift, and Turkey Bolton creeks) remain at risk due to their small population sizes and anthropogenic pressures on remaining habitat. Natural resource managers now have the evolutionary information to guide recovery actions within and among drainages throughout the range of the Okaloosa darter.




Similar content being viewed by others
References
Ayache NC, Near TJ (2009) The utility of morphological data in resolving phylogenetic relationships of darters as exemplified with Etheostoma (Teleostei: Percidae). Bull Peabody Mus Nat Hist 50:327–346
Beneteau CL, Mandrak NE, Heath DD (2007) Characterization of eight polymorphic microsatellite DNA markers for the greenside darter, Etheostoma blennioides (Percidae). Mol Ecol Notes 7:641–643
Burkhead NM, Jelks HL, Jordan F, Weaver DC, Williams JD (1994) The comparative ecology of Okaloosa (Etheostoma okaloosae) and brown (E. edwini) in Boggy and Rocky Bayou stream systems, Choctawhatchee Bay, Florida. Final Report to Eglin Air Force Base, Florida
Castric V, Bonney F, Bernatchez L (2001) Landscape structure and hierarchical genetic diversity in the brook charr, Salvelinus fontinalis. Evolution 55:1016–1028
Champagne CE, Austin JD, Jelks HL, Jordan F (2008) Effects of fin clipping on survival and position-holding behavior of brown darters, Etheostoma edwini. Copeia 2008:917–920
Charlesworth B, Morgan MT, Charlesworth D (1993) The effect of deleterious mutations on neutral molecular variation. Genetics 134:1289–1303
Chen C, Durand E, Forbes F, François O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1660
Coulon A, Fitzpatrick JW, Bowman R, Stith BM, Makarewich CA, Stenzler LM, Lovette IJ (2008) Congruent population structure inferred from dispersal behaviour and intensive genetic surveys of the threatened Florida Scrub-Jay (Aphelocoma cœrulescens). Mol Ecol 17:1685–1701
Crandall KA (1994) Intraspecific cladogram estimation: accuracy at higher levels of divergence. Syst Biol 43:222–235
Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK (2000) Considering evolutionary processes in conservation biology. Trends Ecol Evol 15:290–295
DeSalle R, Amato G (2004) The expansion of conservation genetics. Nat Rev Genet 5:702–712
DeWoody JA, Fletcher DE, Wilkins SD, Avise JC (2000) Parentage and nest guarding in the tessellated darter (Etheostoma olmstedi) assayed by microsatellite markers (Perciformes: Percidae). Copeia 2000:740–747
Dorazio RM, Mukherjee B, Zhang L, Ghosh M, Jelks HL, Jordan F (2008) Modeling unobserved sources of heterogeneity in animal abundance using a Dirichlet process prior. Biometrics 64:635–644
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ewens WJ (1972) The sampling theory of selectively neutral alleles. Theor Popul Biol 3:87–112
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:147–150
Fraser DJ, Bernatchez L (2001) Adaptive evolutionary conservation: towards a unified concept for defining conservation units. Mol Ecol 10:2741–2752
Gabel JM, Dakin EE, Freeman BJ, Porter BA (2008) Isolation and identification of eight microsatellite loci in the Cherokee darter (Etheostoma scotti) and their variability in other members of the genera Etheostoma, Ammocrypta, and Percina. Mol Ecol Resour 8:149–151
Gaggiotti O, Couvet D (2004) Genetic structure in heterogenous environments. In: Ferrière R, Dieckmann U, Couvet D (eds) Evolutionary conservation biology. Cambridge studies in adaptive dynamics. Cambridge University Press, Cambridge, UK, pp 229–243
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodalityin analyses of population structure. Bioinformatics 23:1801–1806
Jordan F, Jelks HL (2009) Population monitoring of the Endangered Okaloosa Darter. Final Report to Eglin Air Force Base, Florida
Jordan F, Jelks HL, Bortone S, Dorazio RM (2008) Comparison of visual survey and seining methods for a benthic stream fish. Environ Biol Fish 81:313–319
Lang NJ, Mayden RL (2007) Systematics of the subgenus Oligocephalus (Teleostei: Percidae: Etheostoma) with complete subgeneric sampling of the genus Etheostoma. Mol Phylogen Evol 43:605–615
Ludwig HR, Leitch JA (1996) Interbasin transfer of aquatic biota via anglers bait buckets. Fisheries 21:14–18
Maguire TL, Peakall R, Saenger P (2002) Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor Appl Gen 104:388–398
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotypic data. Genetics 155:945–959
Rousset P (2008) Genepop’007: a complete re-implementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Schumm SA, Boyd KF, Wolff CG, Spitz WJ (1995) A ground water sapping landscape in the Florida Panhandle. Geomorphology 12:281–297
Song CB, Near TJ, Page LM (1998) Phylogenetic relations among percid fishes as inferred from mitochondrial cytochrome b DNA sequence data. Mol Phylogen Evol 10:343–353
Switzer JF, Welsh SA, King TL (2008) Microsatellite DNA primers for the candy darter, Etheostoma osburni and variegate darter, Etheostoma variatum, and cross-species amplification in other darters (Percidae). Mol Ecol Resour 8:335–338
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis. Genetics 123:585–595
Tajima F (1993) Measurement of DNA polymorphism. In: Takahata N, Clark AG (eds) Mechanisms of molecular evolution. Sinauer Associates Inc, Sunderland, pp 37–59
Templeton AR, Crandall KA, Sin CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonucleases mapping and DNA sequence data. III. Cladogram estimation. Genetics 13:597–601
Tonnis BD (2006) Microsatellite DNA markers for the rainbow darter, Etheostoma caeruleum (Percidae), and their potential utility for other darter species. Mol Ecol Notes 6:230–232
USFWS (1981) Okaloosa darter (Etheostoma okaloosae) recovery plan. U.S. Fish and Wildlife Service, Southeast Region
USFWS (1998) Okaloosa darter (Etheostoma okaloosae) recovery plan (revised). U.S. Fish and Wildlife Service, Southeast Region
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Waples RS (1991) Pacific Salmon, Oncorhynchus spp. and the definition of ‘species’ under the endangered species act. Mar Fish Rev 53:11–22
Waples RS (1995) Evolutionarily significant units and the conservation of biological diversity under the Endangered Species Act. In: Nielsen JL, Powers GA (eds) Evolution and the aquatic ecosystem: defining unique units in population conservation. American Fisheries Society, Bethesda, pp 8–27
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Watterson A, Guess HA (1977) Is the most frequent allele the oldest? Theor Popul Biol 11:141–160
Acknowledgments
We thank B. Garner for her assistance with genotyping. L. Jelks greatly improved the readability of this manuscript. Funding for this research was provided by the Florida Fish and Wildlife Conservation Commission Nongame Grant Program and the U.S. Department of Defense. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the U.S. Fish and Wildlife Service. Mention of trade names or commercial products does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Austin, J.D., Jelks, H.L., Tate, B. et al. Population genetic structure and conservation genetics of threatened Okaloosa darters (Etheostoma okaloosae). Conserv Genet 12, 981–989 (2011). https://doi.org/10.1007/s10592-011-0201-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0201-5