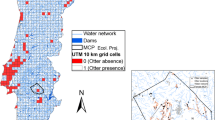
Abstract
Successful conservation and management of rare and elusive species requires reliable estimates of population size, but acquisition of such data is often challenging. We compare the two most frequently used methods of assessing abundance of Eurasian otter (Lutra lutra) populations, noninvasive genetic sampling (NGS) based on genotyping of faeces and field surveys using snow tracking. In a 100-km2 oligotrophic otter habitat with linear water bodies, both methods yielded very similar estimates (10–12 individuals). However, in a 100-km2 fishpond area, consisting of a complex network of rivers, fishponds, channels and marshes, genotyping of faeces revealed the presence of a higher number of individuals (46–50 genotypes) than the snow survey (38 individuals). NGS data analysed by capture-mark-recapture (CMR)-based software CAPWIRE provided even higher estimates, being twice the number assessed through snow tracking (76–81 individuals, CI95% = 49–96 and 55–89). Our results suggest that the performance of both NGS and snow tracking is comparable in simple linear habitats, but in complex habitats with very high otter density a combination of genetic and field methods, or CMR analysis using genetic data, is recommended. We emphasise that to obtain reliable estimates using NGS it is necessary to follow strict protocols for detection and elimination of genotyping errors. Based on a literature review and our experience, we suggest improvements that may increase the success rate and efficiency of NGS for otters.
Similar content being viewed by others
References
Adams JR, Waits LP (2007) An efficient method for screening faecal DNA genotypes and detecting new individuals and hybrids in the red wolf (Canis rufus) experimental population area. Conserv Genet 8:123–131. doi:10.1007/s10592-006-9154-5
Adams JR, Lucash C, Schutte L, Waits LP (2007) Locating hybrid individuals in the red wolf (Canis rufus) experimental population area using a spatially targeted sampling strategy and faecal DNA genotyping. Mol Ecol 16:1823–1834. doi:10.1111/j.1365-294X.2007.03270.x
Arnemo JM, Ahlqvist P, Andersen R, Berntsen F, Ericsson G, Odden J, Brunberg S, Segerström P, Swenson JE (2006) Risk of capture-related mortality in large free-ranging mammals: experiences from Scandinavia. Wildl Biol 12:109–113. doi:10.2981/0909-6396(2006)12[109:ROCMIL]2.0.CO;2
Arrendal J, Vilà C, Björklund M (2007) Reliability of noninvasive genetic census of otters compared to field censuses. Conserv Genet 8:1097–1107. doi:10.1007/s10592-006-9266-y
Bellemain E, Swenson JE, Tallmon D, Brunberg S, Taberlet P (2005) Estimating population size of elusive animals with DNA from hunter-collected feces: four methods for brown bears. Conserv Biol 19:150–161. doi:10.1111/j.1523-1739.2005.00549.x
Bellemain E, Nawaz MA, Valentini A, Swenson JE, Taberlet P (2007) Genetic tracking of the brown bear in northern Pakistan and implications for conservation. Biol Conserv 134:537–547. doi:10.1016/j.biocon.2006.09.004
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273. doi:10.1111/j.1365-294X.2004.02346.x
Broquet T, Petit E (2004) Quantifying genotyping errors in noninvasive population genetics. Mol Ecol 13:3601–3608. doi:10.1111/j.1365-294X.2004.02352.x
Buckland ST, Anderson DR, Burnham KP, Laake JL (1993) Distance sampling: estimating abundance of biological populations. Chapman and Hall, New York
Chanin P (1985) The natural history of otters. Facts On File Inc., New York
Chanin P (2003) Monitoring the otter Lutra lutra. Conserving Natura 2000 Rivers Monitoring Series No. 10, English Nature, Peterborough
Conroy JWH, Chanin PRF (2002) The status of the Eurasian otter (Lutra lutra). In: Dulfer R, Conroy JWH, Nel J, Gutleb AC (eds) Proceedings VIIth international otter colloquium: otter conservation—an example for a sustainable use of wetlands. IUCN Otter Spec. Group Bull 19A/2002, pp 24–48
Conroy JWH, French DD (1987) The use of spraints to monitor populations of otters (Lutra lutra L.). Symp Zool Soc Lond 58:247–262
Coxon K, Chanin P, Dallas J, Sykes T (1999) The use of DNA fingerprinting to study population dynamics of otters (Lutra lutra) in Southern Britain: a feasibility study. R&D Technical Report W202, Environment Agency, Bristol, UK
Creel S, Spong G, Sands JL, Rotella J, Zeigle J, Joe L, Murphy KM, Smith D (2003) Population size estimation in Yellowstone wolves with error-prone noninvasive microsatellite genotypes. Mol Ecol 12:2003–2009. doi:10.1046/j.1365-294X.2003.01868.x
Cutler TL, Swann DE (1999) Using remote photography in wildlife ecology: a review. Wildl Soc Bull 27:571–581
Dallas JF, Piertney SB (1998) Microsatellite primers for the Eurasian otter. Mol Ecol 7:1248–1251
Dallas JF, Bacon PJ, Carss DN, Conroy JWH, Green R, Jefferies DJ, Kruuk H, Marshall F, Piertney SB, Racey PA (1999) Genetic diversity in the Eurasian otter, Lutra lutra, in Scotland. Evidence from microsatellite polymorphism. Biol J Linn Soc Lond 68:73–86. doi:10.1111/j.1095-8312.1999.tb01159.x
Dallas JF, Carss DN, Marshall F, Koepfli K-P, Kruuk H, Piertney SB, Bacon PJ (2000) Sex identification of the Eurasian otter Lutra lutra by PCR typing spraints. Conserv Genet 1:181–183. doi:10.1023/A:1026551510861
Dallas JF, Coxon KE, Sykes T, Chanin PRF, Marshall F, Carss DN, Bacon PJ, Piertney SB, Racey PA (2003) Similar estimates of population genetic composition and sex ratio derived from carcasses and faeces of Eurasian otter Lutra lutra. Mol Ecol 12:275–282. doi:10.1046/j.1365-294X.2003.01712.x
Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples – a case study on DNA in faeces. Front Zool 3:11. doi:10.1186/1742-9994-3-11
Dulfer R, Foerster K, Roche K (1996) Habitat use, home range and behaviour. In: Dulfer R, Roche K (eds) First phase management plan for otters in Třeboň Biosphere Reserve. Nature and Environment 93, Council of Europe Publishing, 1998, pp 24–33
Eggert LS, Eggert JA, Woodruff DS (2003) Estimating population sizes for elusive animals: the forest elephants of Kakum National Park, Ghana. Mol Ecol 12:1389–1402. doi:10.1046/j.1365-294X.2003.01822.x
Erlinge S (1967) Home range of the otter Lutra lutra L. in Southern Sweden. Oikos 18:186–209. doi:10.2307/3565098
Fernando P, Vidya TNC, Rajapakse C, Dangolla A, Melnick DJ (2003) Reliable noninvasive genotyping: fantasy or reality? J Hered 94:115–123. doi:10.1093/jhered/esg022
Ferrando A, Lecis R, Domingo-Roura X, Ponsà M (2008) Genetic diversity and individual identification of reintroduced otters (Lutra lutra) in north-eastern Spain by DNA genotyping of spraints. Conserv Genet 9:129–139. doi:10.1007/s10592-007-9315-1
Flagstad Ø, Hedmark E, Landa A, Brøseth H, Persson J, Anderson R, Segerström P, Ellegren H (2004) Colonization history and noninvasive monitoring of a reestablished wolverine population. Conserv Biol 18:676–688. doi:10.1111/j.1523-1739.2004.00034.x
Foster-Turley P, Macdonald S, Mason C (1990) Otters. An action plan for their conservation. International Union for Conservation of Nature and Natural Resources, Gland
Frantz AC, Pope LC, Carpenter PJ, Roper TJ, Wilson GJ, Delahay RJ, Burke T (2003) Reliable microsatellite genotyping of the Eurasian badger (Meles meles) using faecal DNA. Mol Ecol 12:1649–1661. doi:10.1046/j.1365-294X.2003.01848.x
Garcia de Leaniz C, Forman DW, Davies S, Thomson A (2006) Non-intrusive monitoring of otters (Lutra lutra) using infrared technology. J Zool (Lond) 270:577–584. doi:10.1111/j.1469-7998.2006.00124.x
Garcia DM, Marmontel M, Rosas FW, Santos FR (2007) Conservation genetics of the giant otter (Pteronura brasiliensis (Zimmerman, 1780)) (Carnivora, Mustelidae). Braz J Biol 67:819–827
Gese EM (2001) Monitoring of terrestrial carnivore populations. In: Gittlemann JL, Funk SM, Macdonald D, Wayne RK (eds) Carnivore conservation (conservation biology 5). Cambridge University Press, Cambridge, pp 372–396
Goossens B, Setchell JM, James SS, Funk SM, Chikhi L, Abulani A, Ancrenaz M, Lackman-Ancrenaz I, Bruford MW (2006) Philopatry and reproductive success in Bornean orang-utans (Pongo pygmaeus). Mol Ecol 15:2577–2588. doi:10.1111/j.1365-294X.2006.02952.x
Guter A, Dolev A, Saltz D, Kronfeld-Schor N (2008) Using videotaping to validate the use of spraints as an index of Eurasian otter (Lutra lutra) activity. Ecol Indic 8:462–465. doi:10.1016/j.ecolind.2007.04.009
Hájková P, Bryja J, Zemanová B, Hájek B, Roche K, Zima J (2005) Conservation genetics of Eurasian otters (Lutra lutra): non-invasive genetic sampling and microsatellite DNA variability. Book of abstracts. XIX Annual Meeting of the Society for Conservation Biology, Brasília, Brazil, 15–19 July 2005, p 92
Hájková P, Zemanová B, Bryja J, Hájek B, Roche K, Tkadlec E, Zima J (2006) Factors affecting success of PCR amplification of microsatellite loci from otter faeces. Mol Ecol Notes 6:559–562. doi:10.1111/j.1471-8286.2006.01269.x
Hájková P, Pertoldi C, Zemanová B, Roche K, Hájek B, Bryja J, Zima J (2007) Genetic structure and evidence for recent population decline in Eurasian otter populations in the Czech and Slovak Republics: implications for conservation. J Zool (Lond) 272:1–9. doi:10.1111/j.1469-7998.2006.00259.x
Hansen MM, Jacobsen L (1999) Identification of mustelid species: otter (Lutra lutra), American mink (Mustela vison) and polecat (Mustela putorius), by analysis of DNA from faecal samples. J Zool (Lond) 247:177–181. doi:10.1111/j.1469-7998.1999.tb00981.x
Hansen H, Ben-David M, McDonald DB (2008) Effects of genotyping protocols on success and errors in identifying individual river otters (Lontra canadensis) from their faeces. Mol Ecol Res 8:282–289. doi:10.1111/j.1471-8286.2007.01992.x
Hedmark E, Ellegren H (2006) A test of the multiplex pre-amplification approach in microsatellite genotyping of wolverine faecal DNA. Conserv Genet 7:289–293. doi:10.1007/s10592-005-9000-1
Hedmark E, Ellegren H (2007) DNA-based monitoring of two newly founded Scandinavian wolverine populations. Conserv Genet 8:843–852. doi:10.1007/s10592-006-9231-9
Hung C-M, Li S-H, Lee L-L (2004) Faecal DNA typing to determine the abundance and spatial organisation of otters (Lutra lutra) along two stream systems in Kinmen. Anim Conserv 7:301–311. doi:10.1017/S1367943004001453
Jansman HAH, Chanin PRF, Dallas JF (2001) Monitoring otter populations by DNA typing of spraints. IUCN Otter Spec Group Bull 18:12–19
Janssens X (2006) Monitoring and predicting elusive species colonisation. Application to the otter in the Cévennes National Park (France). PhD Thesis, Université catholique de Louvain, Louvain-la-Neuve, Belgium
Janssens X, Fontaine MC, Michaux JR, Libois R, de Kermabon J, Defourny P, Baret PV (2008) Genetic pattern of the recent recovery of European otters in southern France. Ecography 31:176–186. doi:10.1111/j.0906-7590.2008.4936.x
Kalinowski ST, Taper ML, Creel S (2006) Using DNA from non-invasive samples to identify individuals and census populations: an evidential approach tolerant of genotyping errors. Conserv Genet 7:319–329. doi:10.1007/s10592-005-9024-6
Kalz B, Jewgenow K, Fickel J (2006) Structure of an otter (Lutra lutra) population in Germany – results of DNA and hormone analyses from faecal samples. Mamm Biol 71:321–335. doi:10.1016/j.mambio.2006.02.010
Karanth KU (1995) Estimating tiger Panthera tigris populations from camera-trap data using capture-recapture models. Biol Conserv 71:333–338. doi:10.1016/0006-3207(94)00057-W
Kendall KC, Metzgar LH, Patterson DA, Steele BM (1992) Power of sign survey to monitor population trends. Ecol Appl 2:422–430. doi:10.2307/1941877
Koepfli K-P, Kanchanasaka B, Sasaki H, Jacques H, Louie KDY, Hoai T, Dang NX, Geffen E, Gutleb A, Han S, Heggberget TM, LaFontaine L, Lee H, Melisch R, Ruiz-Olmo J, Santos-Reis M, Sidorovich VE, Stubbe M, Wayne RK (2008) Establishing the foundation for an applied molecular taxonomy of otters in Southeast Asia. Conserv Genet 9:1589–1604. doi:10.1007/s10592-007-9498-5
Kohn MH, Wayne RK (1997) Facts from feces revisited. Trends Ecol Evol 12:223–227. doi:10.1016/S0169-5347(97)01050-1
Kranz A (2000) Otters (Lutra lutra) increasing in Central Europe: from the threat of extinction to locally perceived overpopulation? Mammalia 64:357–368
Kranz A, Knollseisen M (1998) How many otters live ‘here’? A discussion about counting otters. BOKU-Rep Wildl Res Game Manage 14:120–125
Kruuk H (1992) Scent marking by otters (Lutra lutra): signalling the use of resources. Behav Ecol 3:133–140. doi:10.1093/beheco/3.2.133
Kruuk H (1995) Wild otters. Predation and populations. Oxford University Press, Oxford
Kruuk H (2006) Otters: ecology, behaviour, and conservation. Oxford University Press, New York
Kruuk H, Moorhouse A, Conroy JWH, Durbin L, Frear S (1989) An estimate of numbers and habitat preference of otters Lutra lutra in Shetland, UK. Biol Conserv 49:241–254. doi:10.1016/0006-3207(89)90046-3
Kruuk H, Carrs DN, Conroy JWH, Durbin L (1993) Otter (Lutra lutra L.) numbers and fish productivity in rivers in N.E. Scotland. Symp Zool Soc Lond 65:171–191
Lampa S, Gruber B, Henle K, Hoehn M (2008) An optimisation approach to increase DNA amplification success of otter faeces. Conserv Genet 9:201–210. doi:10.1007/s10592-007-9328-9
Lanszki J, Hidas A, Szentes K, Révay T, Lehoczky S, Weiss S (2008) Relative spraint density and genetic structure of otter (Lutra lutra) along the Drava River in Hungary. Mamm Biol 73:40–47. doi:10.1016/j.mambio.2007.08.005
Lucchini V, Fabri E, Marucco F, Ricci S, Boitani L, Randi E (2002) Noninvasive molecular tracking of colonizing wolf (Canis lupus) packs in the western Italian Alps. Mol Ecol 11:857–868. doi:10.1046/j.1365-294X.2002.01489.x
Mason CF, Macdonald SM (2004) Growth in otter (Lutra lutra) populations in the UK as shown by long-term monitoring. Ambio 33:148–152. doi:10.1639/0044-7447(2004)033[0148:GIOLLP]2.0.CO;2
Maudet C, Luikart G, Dubray D, von Hardenberg A, Taberlet P (2004) Low genotyping error rates in wild ungulate faeces sampled in winter. Mol Ecol Notes 4:772–775. doi:10.1111/j.1471-8286.2004.00787.x
McDonald LL (2004) Sampling rare populations. In: Thompson WL (ed) Sampling rare or elusive species: concepts, designs, and techniques for estimating population parameters. Island Press, Washington, DC, pp 11–42
McKelvey KS, Schwartz MK (2004) Genetic errors associated with population estimation using non-invasive molecular tagging: problems and new solutions. J Wildl Manage 68:439–448. doi:10.2193/0022-541X(2004)068[0439:GEAWPE]2.0.CO;2
McKelvey KS, Schwartz MK (2005) DROPOUT: a program to identify problem loci and samples for noninvasive genetic samples in a capture-mark-recapture framework. Mol Ecol Notes 5:716–718. doi:10.1111/j.1471-8286.2005.01038.x
McKelvey KS, von Kienast J, Aubry KB, Koehler GM, Maletzke BT, Squires JR, Lindquist EL, Loch S, Schwartz MK (2006) DNA analysis of hair and scat collected along snow tracks to document the presence of Canada lynx. Wildl Soc Bull 34:451–455. doi:10.2193/0091-7648(2006)34[451:DAOHAS]2.0.CO;2
Miller CR, Joyce P, Waits LP (2005) A new method for estimating the size of small populations from genetic mark-recapture data. Mol Ecol 14:1991–2005. doi:10.1111/j.1365-294X.2005.02577.x
Mills LS, Citta JJ, Lair KP, Schwartz MK, Tallmon DA (2000) Estimating animal abundance using noninvasive DNA sampling: promise and pitfalls. Ecol Appl 10:283–294. doi:10.1890/1051-0761(2000)010[0283:EAAUND]2.0.CO;2
Morin PA, Chambers KE, Boesch C, Vigilant L (2001) Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol Ecol 10:1835–1844. doi:10.1046/j.0962-1083.2001.01308.x
Murphy MA, Kendall KC, Robinson A, Waits LP (2007) The impact of time and field conditions on brown bear (Ursus arctos) faecal DNA amplification. Conserv Genet 8:1219–1224. doi:10.1007/s10592-006-9264-0
Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L (2004) Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Mol Ecol 13:2089–2094. doi:10.1111/j.1365-294X.2004.02207.x
Paetkau D (2003) An empirical exploration of data quality in DNA-based population inventories. Mol Ecol 12:1375–1387. doi:10.1046/j.1365-294X.2003.01820.x
Paetkau C, Strobeck C (1994) Microsatellite analysis of genetic variation in black bear populations. Mol Ecol 3:489–495. doi:10.1111/j.1365-294X.1994.tb00127.x
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. doi:10.1111/j.1471-8286.2005.01155.x
Petit E, Valière N (2006) Estimating population size with noninvasive capture-mark-recapture data. Conserv Biol 20:1062–1073
Piggott MP, Bellemain E, Taberlet P, Taylor AC (2004) A multiplex pre-amplification method that significantly improves microsatellite amplification and error rates for faecal DNA in limiting conditions. Conserv Genet 5:417–420. doi:10.1023/B:COGE.0000031138.67958.44
Piggott MP, Banks SC, Stone N, Banffy C, Taylor AC (2006) Estimating population size of endangered brush-tailed rockwallaby (Petrogale penicillata) colonies using faecal DNA. Mol Ecol 15:81–91. doi:10.1111/j.1365-294X.2005.02783.x
Poledník L, Poledníková K, Hlaváč V, Beran V (2008) Winter census of otters on six sites in the Czech Republic. Bull Vydra 14:11–21 (in Czech with English summary)
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6:847–859. doi:10.1038/nrg1707
Prigioni C, Remonti L, Balestrieri A (2006a) Otter Lutra lutra movements assessed by genotyped spraints in Southern Italy. Hystrix It J Mamm 17:91–96 (ns)
Prigioni C, Remonti L, Balestrieri A, Sgrosso S, Priore G, Mucci N, Randi E (2006b) Estimation of European otter (Lutra lutra) population size by fecal DNA typing in Southern Italy. J Mammal 87:855–858. doi:10.1644/05-MAMM-A-294R1.1
Prugh LR, Ritland CE, Arthur SM, Krebs CJ (2005) Monitoring coyote population dynamics by genotyping faeces. Mol Ecol 14:1585–1596. doi:10.1111/j.1365-294X.2005.02533.x
Puechmaille SJ, Petit EJ (2007) Empirical evaluation of non-invasive capture-mark-recapture estimation of population size based on a single sampling session. J Appl Ecol 44:843–852. doi:10.1111/j.1365-2664.2007.01321.x
Reuther C, Dolch D, Green R, Jahrl J, Jefferies D, Krekemeyer A, Kucerova M, Madsen AB, Romanowski J, Roche K, Ruiz-Olmo J, Teubner J, Trindade A (2000) Surveying and monitoring distribution and population trends of the Eurasian otter (Lutra lutra): guidelines and evaluation of the standard method for surveys as recommended by the European Section of the IUCN/SSC Otter Specialist Group. Habitat 12, Hankensbüttel, Germany
Roche K (2001) Sprainting behaviour, diet, and foraging strategy of stters (Lutra lutra L.) in the Třeboň Biosphere Reserve (Czech Republic). PhD thesis, Institute of Vertebrate Biology, Academy of Sciences of the Czech Republic, Brno, Czech Republic
Roche K, Roche M (2004) Calculating otter (Lutra lutra) numbers in the Třeboň Biosphere Reserve using snow survey data. In: Roche K (ed) Scientific report of the Czech Otter Project 1998–2004. Czech Otter Foundation Fund, Třeboň, pp 132–145
Roon DA, Waits LP, Kendall KC (2005) A simulation test of the effectiveness of several methods for error-checking non-invasive genetic data. Anim Conserv 8:203–215. doi:10.1017/S1367943005001976
Ruiz-Olmo J, Saavedra D, Jiménez J (2001) Testing the surveys and visual and track censuses of Eurasian otters (Lutra lutra). J Zool (Lond) 253:359–369. doi:10.1017/S0952836901000334
Santini A, Lucchini V, Fabbri E, Randi E (2007) Ageing and environmental factors affect PCR success in wolf (Canis lupus) excremental DNA samples. Mol Ecol Notes 7:955–961. doi:10.1111/j.1471-8286.2007.01829.x
Schütz KE, Ågren E, Amundin M, Röken B, Palme R, Mörner T (2006) Behavioral and physiological responses of trap-induced stress in European badgers. J Wildl Manage 70:884–891. doi:10.2193/0022-541X(2006)70[884:BAPROT]2.0.CO;2
Schwartz MK, Pilgrim KL, McKelvey KS, Rivera PT, Ruggiero LF (2007) DNA markers for identifying individual snowshoe hares using field-collected pellets. Northwest Sci 81:316–322
Seber GAF (1982) The estimation of animal abundance, 2nd edn. Macmillan, New York
Sidorovich VE (1997) Mustelids in Belarus—evolutionary ecology, demography and interspecific relationships. Zolotoy Uley, Minsk
Sidorovich VE, Jedrezejewska B, Jedrezejewski W (1996) Winter distribution and abundance of mustelids and beavers in the river valleys of Bialowieza Primeval Forest. Acta Theriol (Warsz) 41:155–170
Šimek L (1997) First estimate of numbers of the otter in the Trebon Biosphere reserve. In: Toman A, Hlavac V (eds) Proceedings 14th Mustelid Colloquium Czech Republic 1995, Praha, pp 81–87
Smallwood KS, Fitzhugh EL (1993) A rigorous technique for identifying individual mountain lions (Felis concolor) by their tracks. Biol Conserv 65:51–59. doi:10.1016/0006-3207(93)90196-8
Smith DA, Ralls K, Hurt A, Adams B, Parker M, Maldonado JE (2006) Assessing reliability of microsatellite genotypes from kit fox faecal samples using genetic and GIS analyses. Mol Ecol 15:387–406. doi:10.1111/j.1365-294X.2005.02841.x
Solberg KH, Bellemain E, Drageset O-M, Taberlet P, Swenson JE (2006) An evaluation of field and non-invasive genetic methods to estimate brown bear (Ursus arctos) population size. Biol Conserv 128:158–168. doi:10.1016/j.biocon.2005.09.025
Sulkava R (2006) Ecology of the otter (Lutra lutra) in Central Finland and methods for estimating the densities of populations. PhD thesis, University of Joensuu, Finland
Sulkava R (2007) Snow tracking: a relevant method for estimating otter Lutra lutra populations. Wildl Biol 13:208–218. doi:10.2981/0909-6396(2007)13[208:STARMF]2.0.CO;2
Taberlet P, Luikart G (1999) Non-invasive genetic sampling and individual identification. Biol J Linn Soc 68:41–55. doi:10.1111/j.1095-8312.1999.tb01157.x
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24:3189–3194. doi:10.1093/nar/24.16.3189
Taberlet P, Waits LP, Luikart G (1999) Noninvasive genetic sampling: look before you leap. Trends Ecol Evol 14:323–327. doi:10.1016/S0169-5347(99)01637-7
Trinca CS, Waldemarin HF, Eizirik E (2007) Genetic diversity of the Neotropical otter (Lontra longicaudis Olfers, 1818) in Southern and Southeastern Brazil. Braz J Biol 64:813–818
Valière N (2002) GIMLET: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2:377–379. doi:10.1046/j.1471-8286.2002.00228.x
Valière N, Berthier P, Mouchiroud D, Pontier D (2002) GEMINI: software for testing the effects of genotyping errors and multitubes approach for individual identification. Mol Ecol Notes 2:83–86
Valière N, Bonenfant C, Toïgo C, Luikart G, Gaillard J-M, Klein F (2007) Importance of a pilot study for non-invasive genetic sampling: genotyping errors and population size estimation in red deer. Conserv Genet 8:69–78. doi:10.1007/s10592-006-9149-2
Waits JL, Leberg PL (2000) Biases associated with population estimation using molecular tagging. Anim Conserv 3:191–199. doi:10.1111/j.1469-1795.2000.tb00103.x
Waits LP, Paetkau D (2005) Noninvasive genetic sampling tools for wildlife biologists: a review of applications and recommendations for accurate data collection. J Wildl Manage 69:1419–1433. doi:10.2193/0022-541X(2005)69[1419:NGSTFW]2.0.CO;2
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256. doi:10.1046/j.1365-294X.2001.01185.x
Wandeler P, Smith S, Morin PA, Pettifor RA, Funk SM (2003) Patterns of nuclear DNA degeneration over time – a case study in historic teeth samples. Mol Ecol 12:1087–1093. doi:10.1046/j.1365-294X.2003.01807.x
Wilson GJ, Delahay RJ (2001) A review of methods to estimate the abundance of terrestrial carnivores using field signs and observation. Wildl Res 28:151–164. doi:10.1071/WR00033
Acknowledgements
The authors would like to thank Miroslav Lehocký, Vašek Bartuška and Jana Moravcová for help with collecting spraint samples in the field, and all the colleagues and volunteers that participated in the snow tracking actions. We appreciate the advice and support of Josef Bryja and Jan Zima. The study was supported by the Czech Science Foundation (grant no. 206/03/0757) and the Czech Ministry of Education (grant no. LC06073).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hájková, P., Zemanová, B., Roche, K. et al. An evaluation of field and noninvasive genetic methods for estimating Eurasian otter population size. Conserv Genet 10, 1667–1681 (2009). https://doi.org/10.1007/s10592-008-9745-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-008-9745-4