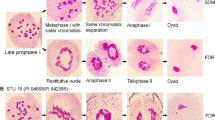
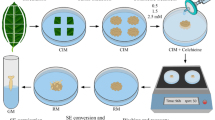
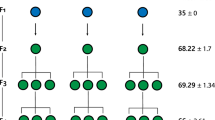
Abstract
Genome instability is observed in several species hybrids. We studied the mechanisms underlying the genome instability in hexaploid hybrids of Napier grass (Pennisetum purpureum R.) and pearl millet (Pennisetum glaucum L.) using a combination of different methods. Chromosomes of both parental genomes are lost by micronucleation. Our analysis suggests that genome instability occurs preferentially in meristematic root tissue of hexaploid hybrids, and chromosome elimination is not only caused by centromere inactivation. Likely, beside centromere dysfunction, unrepaired DNA double-strand breaks result in fragmented chromosomes in synthetic hybrids.







Similar content being viewed by others
Abbreviations
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- GISH:
-
Genomic in situ hybridization
- FISH:
-
Fluorescence in situ hybridization
- CENH3:
-
Centromere-specific histone H3
References
Abreu JC, Davide LC, Vander Pereira A, Barbosa S (2006) Mixoploidy in napiergrass x pearl millet hybrids treated with antimitotic agents. Pesqui Agropecu Bras 41:1629–1635
Andrade-Vieira LF, Reis GB, Torres GA, Oliveira AR, Brasileiro-Vidal AC, Vander Pereira A, Davide LC (2013) Biparental chromosome elimination in artificial interspecific hybrids of Pennisetum purpureum and Pennisetum glaucum. Crop Sci 53:1917–1924
Barbosa S, Davide LC, Pereira AV (2003) Cytogenetics of Pennisetum purpureum Schumack x Pennisetum glaucum L. hybrids and their parents. Ciência Agrotecnol 27(1):26–35
Barbosa S, Davide LC, Pereira AV, Abreu JD (2007) Duplicação cromossômica de híbridos triplóides de capim-elefante e milheto. Bragantia 66(3):365–372
Bennett MD, Finch RA, Barclay IR (1976) The time rate and mechanism of chromosome elimination in Hordeum hybrids. Chromosoma 54(2):175–200
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16(7):1679–1691
Borchert T, Eckardt K, Fuchs J, Kruger K, Hohe A (2009) ‘Who’s who’ in two different flower types of Calluna vulgaris (Ericaceae): morphological and molecular analyses of flower organ identity. BMC Plant Biol 9:148
Campos JMS, Davide LC, Salgado CC, Santos FC, Costa PN, Silva PS, Alves CCS, Viccini LF, Pereira AV (2009) In vitro induction of hexaploid plants from triploid hybrids of Pennisetum purpureum and Pennisetum glaucum. Plant Breed 128:101–104
R Development Core Team (2011) R Foundation for Statistical Computing, Vienna, Austria
Dolezel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2:2233–2244
Finch RA (1983) Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma 88(5):386–393
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220(4601):1049–1051
Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Bruss C, Kumlehn J, Matzk F, Houben A (2005) Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell 17:2431–2438
Grazeffe VS, de Freitas Tallarico L, de Sa Pinheiro A, Kawano T, Suzuki MF, Okazaki K, de Braganca Pereira CA, Nakano E (2008) Establishment of the comet assay in the freshwater snail Biomphalaria glabrata (Say, 1818). Mutat Res 654(1):58–63
Gupta SB, Gupta P (1973) Selective somatic elimination of Nicotiana glutinosa chromosomes in the F1 hybrids of N. suaveolens and N. glutinosa. Genetics 73(4):605–612
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007a) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116(3):275–283
Houben A, Demidov D, Caperta AD, Karimi R, Agueci F, Vlasenko L (2007b) Phosphorylation of histone H3 in plants—a dynamic affair. Biochim Biophys Acta 1769(5–6):308–315
Ishii T, Ueda T, Tanaka H, Tsujimoto H (2010) Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: pearl millet chromosome dynamics in hybrid embryo cells. Chromosome Res 18:821–831
Ishii T, Karimi-Ashtiyania R, Houben A (2016) Haploidization via chromosome elimination: means and mechanisms. Annu Rev Plant Biol (in press)
Ishii T, Tanaka H, Eltayeb AE, Tsujimoto H (2013) Wide hybridization between oat and pearl millet belonging to different subfamilies of Poaceae. Plant Reprod 26:25–32
Ishii T, Sunamura N, Matsumoto A, Eltayeb AE, Tsujimoto H (2015) Preferential recruitment of the maternal centromere-specific histone H3 (CENH3) in oat (Avena sativa L.) × pearl millet (Pennisetum glaucum L.) hybrid embryos. Chromosome Res 23:709–718
Jovtchev G, Menke M, Schubert I (2001) The comet assay detects adaptation to MNU-induced DNA damage in barley. Mutat Res 493:95–100
Karimi-Ashtiyania R, Ishii T, Niessen M, Stein N, Heckmann S, Gurushidze M, Banaei-Moghaddam AM, Fuchs J, Schubert V, Koch K, Weiss O, Demidov D, Schmidt K, Kumlehn J, Houben A (2015) Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc Natl Acad Sci U S A 112:11211–11216
Kasha KJ, Kao KN (1970) High frequency haploid production in barley (Hordeum vulgare L.). Nature 225:874–876
Koçyiğit A, Keles H, Selek S, Guzel S, Celik H, Erel O (2005) Increased DNA damage and oxidative stress in patients with cutaneous leishmaniasis. Mutat Res Genet Toxicol Environ Mutagen 585:71–78
Kuppu S, Tan EH, Nguyen H, Rodgers A, Comai L, Chan SWL, Britt AB (2015) Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet 11, e1005494
Laurie DA (1989) The frequency of fertilization in wheat × pearl millet crosses. Genome 32:1063–1067
Leao FF, Davide LC, de Campos JMS, Vander Pereira A, Bustamante FD (2011) Genomic behavior of hybrid combinations between elephant grass and pearl millet. Pesqui Agropecu Bras 46:712–719
Leitch AR, Leitch IJ (2008) Perspective—genomic plasticity and the diversity of polyploid plants. Science 320:481–483
Levy AA, Feldman M (2004) Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol J Linn Soc 82:607–613
Liu M, Li ZY (2007) Genome doubling and chromosome elimination with fragment recombination leading to the formation of Brassica rapa-type plants with genomic alterations in crosses with Orychophragmus violaceus. Genome 50:985–993
Ma XF, Gustafson JP (2005) Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenet Genome Res 109:236–249
Maheshwari S, Tan EH, West A, Franklin FCH, Comai L, Chan SWL (2015) Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet 11, e1004970
Marques de Resende KF, Santos FMC, de Paula CP, Techio VH, Davide LC (2015) Effects of hybridization and polyploidy on the histone H3 phosphorylation at serine 10 (H3S10ph) in Pennisetum spp. Rich. (Poaceae). Aust J Crop Sci 9(5):453–45
Martel E, DeNay D, SiljakYakovlev S, Brown S, Sarr A (1997) Genome size variation and basic chromosome number in pearl millet and fourteen related Pennisetum species. J Hered 88:139–143
Mochida K, Tsujimoto H, Sasakuma T (2004) Confocal analysis of chromosome behavior in wheat x maize zygotes. Genome 47:199–205
Noda K, Kasha KJ (1981) Chromosome elimination in different meristematic regions of hybrids between Hordeum vulgare L, and Hordeum bulbosum L. Jpn J Genet 56:193–204
Pereira AV, Ferreira RP, Passos LP, Freitas VP, Verneque RS, Barra RB, Silva CHP (2000) Variação da qualidade de folhas em capim elefante (Pennisetum purpureum) e híbridos de capim elefante x milheto (P. purpureum x P. glaucum), em função da idade da planta. Ciênc Agrotec 24(2):490–499
Ravi M, Chan SWL (2010) Haploid plants produced by centromere-mediated genome elimination. Nature 464:615–U180
Reis GB, Mesquita AT, Torres GA, Andrade-Vieira LF, Pereira AV, Davide LC (2014) Genomic homeology between Pennisetum purpureum and Pennisetum glaucum (Poaceae). Comp Cytogenet 8:199–209
Riddle NC, Birchler JA (2003) Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet 19:597–600
Sanei M, Pickering R, Kumke K, Nasuda S, Houben A (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A 108:E498–E505
Souza-Sobrinho FD, Pereira AV, Ledo FJD, Botrel MA, Oliveira JSE, Xavier DF (2005) Agronomic evaluation of interespecific hybrids of elephant grass and pearlmillet. Pesqui Agropecu Bras 40:873–880
Tan EH, Henry IM, Ravi M, Bradnam KR, Mandakova T, Marimuthu MPA, Korf I, Lysak MA, Comai L, Chan SWL (2015) Catastrophic chromosomal restructuring during genome elimination in plants. eLife 2015; doi:10.7554/eLife.06516.
Tang ZX, Fu SL, Yan BJ, Zhang HQ, Ren ZL (2012) Unequal chromosome division and inter-genomic translocation occurred in somatic cells of wheat-rye allopolyploid. J Plant Res 125:283–290
Tiwari VK, Rawat N, Neelam K, Kumar S, Randhawa GS, Dhaliwal HS (2010) Random chromosome elimination in synthetic Triticum-Aegilops amphiploids leads to development of a stable partial amphiploid with high grain micro- and macronutrient content and powdery mildew resistance. Genome 53:1053–1065
van Gent DC, Hoeijmakers JHJ, Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2(3):196–206
Wang Z, Yin H, Lv L, Feng YY, Chen SP, Liang JT, Huang Y, Jiang XH, Jiang HW, Bukhari I, Wu LJ, Cooke HJ, Shi QH (2014) Unrepaired DNA damage facilitates elimination of uniparental chromosomes in interspecific hybrid cells. Cell Cycle 13:1345–1356
Acknowledgments
The authors would like to thank the FAPEMIG (Fundação de Amparo a Pesquisa do Estado de Minas Gerais), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Cientifico e Tecnológico), and IPK (Gatersleben) for providing the financial support; EMBRAPA (Brazilian Corporation of Agricultural Research) for the plant material; and Marcel José Palmieri for the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Hans de Jong
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure Suppl. 1
Flow cytometry histograms of analyzed Napier grass, pearl millet and hexaploid hybrids (6X-1, 6X-2, 6X-3, 6X-4 and 6X-5). (DOC 497 kb)
Figure Suppl. 2
Fluorescence in situ hybridization with centromere-specific probes. (A) Napier grass (NG). (B) Pearl millet (PM). Bars = 10 μm. (DOC 304 kb)
ESM 3
(DOC 61 kb)
Rights and permissions
About this article
Cite this article
dos Reis, G.B., Ishii, T., Fuchs, J. et al. Tissue-specific genome instability in synthetic interspecific hybrids of Pennisetum purpureum (Napier grass) and Pennisetum glaucum (pearl millet) is caused by micronucleation. Chromosome Res 24, 285–297 (2016). https://doi.org/10.1007/s10577-016-9521-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-016-9521-0