Abstract
The Malagasy primate family Indriidae comprises three genera with up to 19 species. Cytogenetic and molecular phylogenies of the Indriidae have been performed with special attention to the genus Propithecus. Comparative R-banding and FISH with human paints were applied to karyotypes of representatives of all three genera and confirmed most of the earlier R-banding results. However, additional chromosomal rearrangements were detected. A reticulated and a cladistic phylogeny, the latter including hemiplasies, have been performed. Cladistic analysis of cytogenetic data resulted in a phylogenetic tree revealing (1) monophyly of the family Indriidae, (2) monophyly of the genus Avahi, (3) sister–group relationships between Propithecus diadema and Propithecus edwardsi, and (4) the grouping of the latter with Indri indri, Propithecus verreauxi, and Propithecus tattersalli, and thus suggesting paraphyly of the genus Propithecus. A molecular phylogeny based on complete mitochondrial cytochrome b sequences of 16 species indicated some identical relationships, such as the monophyly of Avahi and the sister–group relationships of the eastern (P. diadema and P. edwardsi) to the western Propithecus species (P. verreauxi, Propithecus coquereli, and P. tattersalli). However, the main difference between the molecular and cytogenetic phylogenies consists in an early divergence of Indri in the molecular phylogeny while in the chromosomal phylogeny it is nested within Propithecus. The similarities and differences between molecular and cytogenetic phylogenies in relation to data on the species’ geographic distributions and mating systems allow us to propose a scenario of the evolution of Indriidae. Chromosomal and molecular processes alone or in combination created a reproductive barrier that was then followed by further speciation processes.
Similar content being viewed by others
Introduction
The island of Madagascar is famous for its high rate of endemic plant and animal species and for its species richness. Around 100 endemic primate species are described (Mittermeier et al. 2010), all of which are descendants from a single ancestral population that colonized the island in the early Eocene (Horvath et al. 2008; Roos et al. 2004). Although knowledge about the diversity and evolution of Madagascar’s primate species has increased in the last decades, detailed information for most taxa is still missing. This is especially true for members of the family Indriidae. While there is consensus that the family contains the three genera, Indri, Propithecus, and Avahi, the number of species to be recognized and phylogenetic relationships among them remains disputed.
Among the three genera, the nocturnal woolly lemurs of the genus Avahi are the smallest taxa (~1 kg). Originally only one species, Avahi laniger (2n = 70), with an eastern and western subspecies was recognized (Schwarz 1931; Petter et al. 1977; Tattersall 1982). Subsequently, both subspecies were elevated to full species, A. laniger and Avahi occidentalis by Rumpler et al. (1990) based on cytogenetic characters. Later, two additional species, Avahi unicolor and Avahi cleesei, were described based on morphological characters by Thalmann and Geissmann (2000, 2005). Recent molecular investigations further increased the number of species to seven (Zaramody et al. 2006) and even to nine (Andriantompohavana et al. 2007; Lei et al. 2008). Although the taxonomic status of several of these forms remains disputed (Zaramody et al. 2006), we follow here the current classification with nine Avahi species (Mittermeier et al. 2010) (for species ranges, see Fig. 1).
Sifakas, genus Propithecus, are diurnal and larger than woolly lemurs. The genus originally contained two species, the smaller Propithecus verreauxi (3–4.5 kg) with four subspecies (P. v. coquereli, P. v. coronatus, P. v. deckeni, P. v. verreauxi) from the western and southern dry forests, and the larger P. diadema (5–8.5 kg) with five subspecies (P. d. candidus, P. d. diadema, P. d. edwardsi, P. d. holomelas, P. d. perrieri) from eastern Madagascar (Hill 1953; Petter et al. 1977). Tattersall (1986), however, regarded P. d. holomelas as synonym of P. d. edwardsi, while Simons (1988) added P. tattersalli as an additional species. In recent classifications, all previous subspecies of P. diadema (Mayor et al. 2004) and P. verreauxi were elevated to distinct species, resulting in a total of nine species (Mittermeier et al. 2010).
For the P. diadema group, cytogenetic investigations distinguished between two karyotypes (2n = 42 for P. diadema, P. candidus, and P. perrieri, and 2n = 44 for P. edwardsi), while all members of the P. verreauxi group carry the same diploid chromosome number of 2n = 48 (Rumpler and Albignac 1979; Rumpler et al. 2004). P. tattersalli shows a diploid chromosome number of 2n = 42 (Rumpler et al. 2004). Based on mitochondrial sequence data, three major clades among sifakas were detected (Razafindraibe et al. 2000; Pastorini et al. 2001; Mayor et al. 2004). The first comprises all members of the P. diadema group, in the second P. tattersalli and P. coquereli cluster together, and the third contains the remaining members of the P. verreauxi group.
In contrast to the former two genera, the genus Indri is monotypic comprising the single species Indri indri (2n = 40). Indris are the largest extant lemurs (6–9.5 kg) and occur only in the central to northern parts of the eastern Malagasy forest with a range extending from the Anosibe an’ala Classified Forest in the south to the Anjanaharibé-Sud Special Reserve in the north (Petter et al. 1977; Tattersall 1982; Powzyk and Thalmann 2003). This species is well characterized by both karyotypic and molecular data (Rumpler et al. 1988; Delpero et al. 2001; Roos et al. 2004).
Although chromosomal (Rumpler et al. 2004) and mitochondrial sequence data (Pastorini et al. 2001; Mayor et al. 2004; Roos et al. 2004; Zaramody et al. 2006) are available for several Indriidae species, all these studies are characterized by an incomplete taxonomic and geographic sampling, especially for Propithecus. Moreover, some taxa have been split into several species within the last years and therefore might have been misidentified in earlier studies. To obtain a more complete picture of the evolution of this enigmatic primate family, information of recently discovered species and samples with known geographic provenance are necessary. Furthermore, Rumpler et al. (2004) proposed that the most parsimonious model for chromosomal evolution in Indriidae would include a phase of reticulation. In light of possible hemiplasic relationships in chromosomal phylogenies, as suggested by Robinson et al. (2008) and Avise and Robinson (2008), it is necessary to explore whether reticulation is still the most parsimonious model.
In our study, we analyze the phylogenetic relationships between the species of the three indriid genera by cladistic methods using cytogenetic data derived from comparative R-banding and fluorescence in situ hybridization (FISH) with human paints. We establish a cladistic phylogeny and compare it with the model of reticulated chromosomal evolution proposed in an earlier study (Rumpler et al. 2004). Furthermore, we establish a molecular phylogeny based on mitochondrial cytochrome b (cyt b) sequence data from all three indriid genera and most of their species, and compare it with the chromosomal phylogenetic patterns. This enables us to propose an evolutionary scenario based on phylogenetic analyses of molecular and cytogenetic data in combination with data on the species’ geographic distributions. This allows us also to refine the distributions of parapatric Propithecus species pairs (P. diadema/P. edwardsi and P. coronatus/P. deckeni) (Curtis et al. 1998; P. Moisson, personal communication).
Methods
Samples
For Propithecus, 55 tissue samples were collected during several field surveys (2002–2008) in different parts of the eastern forests of Madagascar. Samples were collected from Analamera in the north to Ranomafana in the south, and in the western forests from Ampijoroa in the north to Kirindy in the south (Fig. 1, Supplementary Table S1). Animals were caught and anesthetized with a 2 mg/kg injection of ketamine solution (Ketalar® Parke-Davis). Skin biopsies were taken under general anesthesia and frozen in liquid nitrogen after adding DMSO (10%) as cryoprotector. The samples were first kept at the Institute Pasteur of Antananarivo and then sent to the Institute of Embryology of the University of Strasbourg, France. The collection of samples was approved by the respective Malagasy authorities.
For the cytogenetic study, we used, in addition to the Propithecus samples, fibroblast cultures from Avahi species, I. indri, and the two outgroup species Microcebus murinus and Eulemur fulvus which were already present at the cell bank of the Institute of Embryology (Table 1). For the molecular analysis, we used all 55 Propithecus samples, one sample of Indri from the cell bank of the Institute of Embryology, and the respective data previously published by Zaramody et al. (2006) from 49 samples of Avahi. Latter samples were collected in different parts of the eastern Malagasy forests from Antsahaporetiny in the north to Fort Dauphin in the south and in the western forests from Ampijoroa and Bemahara (Fig. 1). No samples were available for A. mooreorum, A. unicolor, and P. candidus.
Cytogenetics
R-banding was performed on three samples of P. diadema from Ampambelezapiko, Ivongo, and Mantadia; four P. edwardsi from Ambodisovoka, Anjamba, and Vohipara; three P. tattersalli from Daraina; one P. verreauxi from Kirindy; one P. coquereli from Ampijoroa; and two P. coronatus from the Zoo of Mulhouse, France (Table 1). From all other Indriidae species, R-banded karyotypes were taken from Rumpler et al. (2004). For 12 samples which did not give enough mitosis to allow R-banding, only Giemsa staining was performed (Table 1).
FISH techniques were applied to the chromosomes of one sample of each of the following nine species: P. verreauxi, P. coronatus, P. coquereli, P. diadema, P. edwardsi, P. tattersalli, I. indri, A. laniger, and A. occidentalis. We tried to use a complete set of human chromosome paints for the different species, but when the repetitive fibroblast cultures of a species gave rise only to a reduced number of slides showing numerous metaphases with well-dispersed chromosomes, we chose not to paint chromosomes with similar R-banding pattern in closely related species. In detail, we used the following paints in the different species: HSA1, HSA12, HSA16, HSA17, and HSA19 for all species; HSA2, HSA3, HSA4, HSA6, HSA11, HSA14, HSA15, and HSA21 for all species except P. edwardsi; HSA5 for all except P. edwardsi and P. diadema; HSA7 and HSA8 for all except P. edwardsi and P. tattersalli; HSA10 for all species except A. occidentalis; HSA13 for all except A. occidentalis and P. diadema; and HSA18, HSA20, and HSA22 for all except P. edwardsi, P. tattersalli, and A. occidentalis. FISH experiments were conducted using commercially available whole human (HSA) chromosome paints (Biosys SA—Cambio, Cambridge, UK) for all autosomes. In situ hybridization was performed as described in the manufacturer’s instructions with some modifications (Vezuli and Rumpler 2000). After incubation for 72 h, slides were washed at 40°C. Biotinylated DNA probes were detected via avidin coupled with fluorescein isothiocyanate (FITC) and visualized without further amplification of the signal. Metaphases were counterstained with propidium iodide or DAPI. The hybridization signals were observed via an epifluorescence Zeiss Axioplan microscope and digital images were taken with a cooled CCD camera (JAI M300) linked to the microscope. As imaging software, Isis3 (MetaSystem GmbH, Germany) was used.
We reconstructed and compared a cladistic phylogeny and a phylogeny containing a reticulate phase. In the cladistic model, changes of the karyotypic structure between the different Indriidae and outgroup species were identified and then coded as binary (presence = 1, absence = 0) characters following the principles described in Dobigny et al. (2004). Only informative, i.e., different character states between outgroup taxa and at least one of the ingroup taxa, and autapomorphic characters were included in the matrix (Supplementary Table S2). As outgroup taxa, we used M. murinus (MMR, 2n = 66) and E. fulvus (EFU, 2n = 60) since both are representatives of two other Malagasy lemur families (Cheirogaleidae and Lemuridae, respectively), and since for both species detailed R- and C-banding (Rumpler and Dutrillaux 1990) as well as painted data are available. The same outgroups were chosen in an earlier study on sportive lemurs (Rumpler et al. 2008). Primary homologies were identified by comparing R-banding patterns and FISH data from the seven cytogenetically different Indriidae species (A. laniger 2n = 70, A. occidentalis 2n = 70, P. tattersalli 2n = 42, P. diadema 2n = 42, P. edwardsi 2n = 44, P. verreauxi 2n = 48, and I. indri 2n = 40) and the two outgroup taxa. Importantly, no a priori polarization (e.g., translocation vs. fission) was assumed and changes were all strictly inferred by outgroup comparison criteria, thus allowing an a posteriori reconstruction of chromosome evolution. We constructed a phylogenetic tree based on the binary coded chromosomal rearrangements with the maximum-parsimony (MP) algorithm where some reversions and/or convergence rearrangements are allowed. The MP tree was calculated via an exhaustive search in PAUP 4.0b10 (Swofford 2000) and robustness of internal branches was verified with 10,000 bootstrap replications.
In a second approach, we considered possible reversions or convergences as rearrangements that existed at an ancestral polymorphic stage which were only retained in some lineages while they were lost in others, thus leading to hemiplasic relationships (Avise and Robinson 2008; Robinson et al. 2008). The absence of such rearrangements, however, does not correspond to the reversion of a fixed rearrangement.
For the reticulate model, we manually reconstructed a hypothetical ancestral karyotype by comparing the karyotypes of the different species according to the method previously used by Dutrillaux (1986) and Rumpler et al. (2004). The respective phylogenetic tree, starting from this ancestral karyotype and leading to the different species, was constructed in the most parsimonious way without convergence or reversion. Consequently, rearrangements which appeared to be reversions or convergences in the cladistic reconstruction have to be exchanged by hybridization between diverging lineages. The question whether a reticulated evolution is a priori more likely to occur than reversions or convergences remains debatable. Indeed, there is clear evidence that convergences and reversions are common in chromosomal evolution. Such cases have been reported for several rodent species (Qumsiyeh et al. 1987; Volobouev et al. 2002; Britton-Davidian et al. 2005) and lemurs (Warter et al. 2005).
Molecular genetics
A molecular phylogeny was reconstructed with sequences of the complete mitochondrial cyt b gene (1,140 bp). Therefore, 38 unique sequences (37 for Avahi, one for Indri) taken from GenBank were combined with 55 newly generated Propithecus sequences (Supplementary Table S1). DNA extraction, PCR amplification, and sequencing followed methods as described in Roos et al. (2004) and Andriaholinirina et al. (2006). Due to the absence of indels, the alignment was easily generated by eye. To exclude the possibility that nuclear pseudogenes (numts) were amplified, all sequences were checked for premature stop codons. The final Indriidae data set included 71 unique haplotypes representing 16 taxa.
Phylogenetic trees were constructed with maximum-likelihood (ML) and Bayesian algorithms using GARLI 0.951 (Zwickl 2006) and MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). As outgroup taxa, we used Lepilemur ruficaudatus, Phaner furcifer, Lemur catta, and Daubentonia madagascariensis, which were all available in GenBank. For both reconstructions, the HKY + I + Γ model was applied as it was chosen as best-fit model under the Akaike Information Criterion by MODELTEST 3.7 (Posada and Crandall 1998). In GARLI, only the model specification settings were adjusted according to the dataset, while all other settings were left at their default value. Support of internal branches was calculated with 500 bootstrap replications. ML majority-rule consensus trees were calculated in PAUP. For Bayesian reconstructions, we used four Markov Chain Monte Carlo chains with the default temperature of 0.1. Four repetitions were run for 10,000,000 generations with tree and parameter sampling occurring every 100 generations. The first 25% of samples were discarded as burnin, leaving 75,001 trees per run. Posterior probabilities for each split and a phylogram with mean branch lengths were calculated from the posterior density of trees.
Results
Cytogenetics
Recurrent signals were obtained for all hybridized chromosome painting probes (Fig. 2). The hybridization quality, however, was much lower than that seen for human chromosomes on the same slide. Among the 16 karyotyped taxa, only seven unique karyotypes were detected (Table 2): A. occidentalis (=A. cleesei), A. laniger (=A. peyrierasi = A. ramanantsoavani = A. meridionalis = A. betsileo), P. diadema (=P. perrieri), P. edwardsi, P. tattersalli, P. verreauxi (=P. coronatus = P. deckeni = P. coquereli), and I. indri. This result confirms and complements earlier findings from Rumpler et al. (1988), Mayor et al. (2004), and Zaramody et al. (2006).
In Table 3, homologous segments between the chromosomes of these karyotypes and the two outgroup taxa as obtained from R-banding techniques are listed. The FISH techniques mostly confirmed earlier results solely based on R-banding techniques (Rumpler et al. 2004), but allowed us to specify more precisely some other rearrangements and expanded results of earlier studies (Table 3).
As a result, we present the following new cytogenetic data: one previously undetermined rearrangement involving three minute chromosomes to give rise to IIN10 has been replaced by one Robertsonian translocation followed by a centromere–telomere one [50] (numbers in squared parentheses refer to rearrangements as listed in Table 4 and are also identical to rearrangements in Figs. 3 and 4). In addition, one undetermined rearrangement giving PTA14 has been replaced by a Robertsonian translocation [49]. ALA3 which was the result of a complex rearrangement including two chromosomes finally appears as a result of three rearrangements [15,16,17]. Two minute chromosomes of Avahi which could not be identified in earlier studies are the result of a centromere–telomere fission [34]. On the other hand, two rearrangements [32] and [33] which were considered as common for P. verreauxi, P. diadema, and Indri and specific for P. tattersalli before (Rumpler et al. 2004), are now considered as a common shift for all Indriidae [32] which has undergone an additional shift [33] in P. tattersalli. The common occurrence of P. tattersalli and I. indri was inferred in earlier studies. The FISH data also confirm the following human chromosome associations 3–21, 14–15, 12–22, which were already found with R-banding and to detect the following associations 7–16, 4–2, 8–15, 4–19, 6–4, but the last was not present in Avahi.
Parsimony reconstruction of chromosomal changes allowing for hemiplasies. Numbering of rearrangements as in Table 4. Hemiplasic rearrangements are indicated by “X”. “X” depicted in a broken square indicates an ancestral polymorphic stage of the respective rearrangement, whereas when depicted in closed squares, these rearrangements have not been retained in the lineage. Numbers in gray circles represent bootstrap values
The comparison of R-banding patterns and FISH data allowed us to identify all autosomal euchromatic homologous chromosomes and chromosomal segments, and to define their presence/absence pattern in the analyzed species (Table 4).
The respective cladistic tree, based on presence/absence data (Supplementary Table S2), is depicted in Fig. 3. In this reconstruction, however, we allowed for hemiplasies (Avise and Robinson 2008; Robinson et al. 2008).
Accordingly, from Microcebus as a starting point, a common trunk bearing three centromere–telomere translocations [1,13] and one inversion [19] gives rise to two branches. One branch leads to E. fulvus which carries one Robertsonian translocation [2], one centromere–telomere translocation [26], and one centromere–telomere fission [9]. The second branch leads to the Indriidae. The Indriidae branch bears three shifts [14,30,32], one Robertsonian fission [20], and one inversion [43]. This branch then bifurcates into two clades. The first leading to Avahi carries three centromere–telomere translocations [16,17,29], four Robertsonian fissions [15,31,34,45], and one centromere–telomere fission [28]. Both A. laniger and A. occidentalis lineages contain each one non-homologous Robertsonian translocation [4 and 10] and A. laniger additionally a gain of heterochromatin [3].
The second branch bears four Robertsonian translocations [11,18,24,41] and two inversions [46,47]. The three rearrangements [11,18,41], common to Propithecus, are absent in Indri, and the rearrangement [22], common for Indri and P. tattersalli, but absent in P. verreauxi, can be regarded as hemiplasic. The respective branch diverges further into two subsequent lineages. The first is characterized by six Robertsonian translocations [6,21,25,36,39,51]. This lineage further diverges into a P. edwardsi and a P. diadema branch, which is defined by an additional Robertsonian translocation [48]. The second branch, bearing one inversion [27] and three Robertsonian translocations [22,38,40], of which one [22] is hemiplasic, splits into a P. verreauxi/Indri branch with one additional Robertsonian translocation [35] and a P. tattersalli lineage with three Robertsonian translocations [7,37,49], one shift [33], and one centromere–telomere translocation [44]. The branch leading to Indri bears a Robertsonian translocation followed by one inversion [8], one Robertsonian translocation [12], one transposition [23], one Robertsonian translocation followed by two centromere–telomere translocations [42], and one Robertsonian translocation followed by a centromere–telomere translocation [50], while the branch leading to P. verreauxi carries one Robertsonian translocation [5].
In total, the 51 associations consist of 56 chromosomal rearrangements which comprise 26 Robertsonian translocations (the hemiplasic rearrangements have only be counted when they occurred on their polymorphic stage), six inversions, four shifts, 11 centromere–telomere translocations, five Robertsonian fissions, two centromere–telomere fissions, one transposition, and one gain of heterochromatin.
The reticulated phylogeny is depicted in Fig. 4. In principal, obtained relationships are the same as in Fig. 3, starting with the consecutive divergences of EFU and Avahi. The branch leading to Indri and Propithecus bears two inversions and one Robertsonian translocation [24,46,47]. The following reticulated evolution thus involves eight rearrangements, among them the four hemiplasic rearrangements [11,18,41,22]: two Robertsonian translocations [38,40] and one inversion [27] shared by Indri, P. verreauxi, and P. tattersalli; three Robertsonian translocations [11,18,41] shared by P. verreauxi, P. tattersalli, and P. diadema; one Robertsonian translocation shared by Indri and P. verreauxi [35]; and one Robertsonian translocation shared by Indri and P. tattersalli [22]. From this network, various branches emerge containing the respective specific rearrangements identical to those of the cladistic phylogeny. The branch leading to Indri bears nine rearrangements: one Robertsonian translocation followed by an inversion [8], one Robertsonian translocation [12], one transposition [23], one Robertsonian translocation followed by two centromere–telomere translocations [42], and one Robertsonian translocation followed by one centromere–telomere translocation [50]. The branch leading to P. verreauxi bears only one Robertsonian translocation [5], whereas the branch leading to P. tattersalli carries three Robertsonian translocations [7,37,49], one centromere–telomere translocation [44], and one shift [33]. The branch to P. edwardsi carries six Robertsonian translocations [6,21,25,36,39,51] and on the P. diadema lineage one additional Robertsonian translocation appears [48]. All 56 rearrangements were necessary to give rise to the tree without hemiplasy, convergence, or reversion.
Molecular genetics
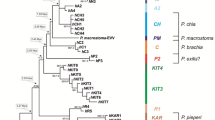
Phylogenetic trees reconstructed on the basis of ML and Bayesian algorithms revealed identical tree topologies (Fig. 5). Although the monophyly of the Indriidae family was well supported, the relationships among the three genera Avahi, Indri, and Propithecus remained unresolved. Within Avahi, we observed a further division into an eastern clade containing A. laniger, A. peyrierasi, A. meridionalis, and A. ramanantsoavani, and a western clade with A. cleesei and A. occidentalis. The Propithecus clade further divides into two subgroups with geographically non-overlapping ranges. The first of these comprises the western populations of the P. verreauxi group and P. tattersalli, and the second the eastern populations of the P. diadema group. The latter further splits into four clades: P. perrieri, P. diadema 1, P. diadema 2, and P. edwardsi. P. perrieri occurs north of the eastern forest in Daraina, P. diadema 2 occurs in an area extending from Zahamena in the northern part till Maromizaha in the central eastern part of the P. diadema range, while P. diadema 1 encompasses the P. diadema 2 range in the North, West, and South (Fig. 1). The Mangoro/Onive Rivers comprise the southern limit of the P. diadema distribution. P. edwardsi contains all populations from the Mangoro/Onive Rivers in the north as far south as Anjamba and Ranomafana. The clade comprising members of the P. verreauxi group and P. tattersalli further diverges into two subclades. The southern subclade consists of P. verreauxi, P. coronatus, P. deckeni, and P. spp., and the northern subclade of P. coquereli and P. tattersalli, thus suggesting paraphyly of the P. verreauxi group. This appears particularly evident for P. deckeni and P. coronatus due to the morphological aspect of some animals (P. spp.) suggesting hybridization.
Discussion
Chromosomal evolution
In the course of the evolution of the Indriidae, the occurrence of several centromere–telomere translocations suggests that during chromosomal evolution new centromeres occurred while on some chromosomes, others were deactivated. These preserved their structural integrity in at least some chromosomes as a result of earlier centromere–telomere translocations. At the level of the obtained resolution, it is likely that the same breakpoints corresponding to active or latent centromeres were repeatedly used during the karyotype restructuring. This would also explain the occurrence of reverse mutations and probably the propensity to chromosomal polymorphisms (Kehrer-Sawatzki and Cooper 2008; Stanyon et al. 2008).
Comparison between cladistic and reticulated trees
In the cladistic tree, after the split between Avahi and the other Indriidae, four Robertsonian translocations [11,18,22,41] appeared as homoplasic. Indeed, three of them are present in all Propithecus but absent in Indri and can be considered either as convergences, the same rearrangements having occurred independently on three branches, or as having undergone reversions on the branch leading to Indri. The latter might be most parsimonious. The fourth rearrangement, present in Indri and P. tattersalli but absent in P. verreauxi, can also be considered as the result of one convergence or reversion. By using the interpretative hypothesis of Avise and Robinson (2008) and Robinson et al. (2008) termed hemiplasy, these four rearrangements are considered as having been ancestrally present on a polymorphic stage and they were only retained in some karyotypes while they disappeared in others. Hemiplasy is more plausible for neutral polymorphism such as Robertsonian translocations which have little or no impact on fertility (Robinson et al. 2008). Indeed, crosses between different species or subspecies of lemurs differing by six and eight rearrangements have been observed in the wild and in captivity, respectively, without reducing the viability of the hybrids (Rumpler and Dutrillaux 1990). A reduction of fertility in male hybrids is nevertheless observed if the Robertsonian translocations involve the same acrocentric chromosomes but in different combinations, so that the resulting metacentric chromosomes show monobrachial homologies. This leads to the formation of meiotic multivalents and to significant gametogenic impairment if the multivalents comprise more than four chromosomes. In females, in contrast, ovogenesis seems not to be disturbed in species studied so far (Rumpler and Dutrillaux 1990). In the chromosomal evolution of the Indriidae, the Robertsonian translocations involved in the polymorphic stage would give rise to multiple independent heterozygosities and none multivalents, and hence, would be no major obstacle for a successful reproduction. Conversely, on the specific branches, the Robertsonian translocations gave rise to chromosomes showing monobrachial homologies which are more selective in terms of speciation.
In the reticulated evolution, eight rearrangements (seven Robertsonian translocations and one inversion) are involved in the reticulated process. Among them the four abovementioned Robertsonian translocations [11,18,22,41] and three [27,38,40] occurring on the common branch to P. verreauxi/P. tattersalli/Indri and one [35] on the branch to P. verreauxi and I. indri. Hybridization during this phase would give rise to seven independent trivalents and a single inversion loop, which would not necessarily reduce fertility of the heterozygotes. Conversely, the rearrangements on the taxon-specific branches to Indri, P. verreauxi, P. tattersalli, and P. edwardsi/P. diadema, which are identical in both phylogenies, will have the same speciation effect. It appears to us that the cladistic phylogeny with four polymorphic (hemiplasic) rearrangements is the most likely and the most similar to the molecular phylogeny. Thus, we will retain it for the comparison between chromosomal and molecular phylogenies.
Chromosomal versus molecular phylogeny
By comparing phylogenetic trees inferred from chromosomal (Fig. 3) and molecular data (Fig. 5), some identical relationships among species and clades became obvious. In both phylogenies, the Avahi clade as well as the sister group relationships of P. diadema/P. edwardsi to P. verreauxi/P. tattersalli are strongly supported. Moreover, a major split within the genus Avahi separating the eastern and the western species is also strongly supported by both phylogenies even if the chromosome characteristics do not allow distinguish between as many species as the molecular data.
However, the following relationships differ among phylogenies: (1) Although phylogenetic relationships among the three Indriidae genera are not resolved, the molecular phylogeny indicates an early emergence of Indri while in the chromosomal phylogenies Indri is nested within Propithecus. (2) Whereas the chromosome data indicate a clade consisting of P. coquereli, P. deckeni, P. coronatus, and P. verreauxi, the mitochondrial phylogeny suggests a closer relationship of P. coquereli to P. tattersalli than to the other taxa of the P. verreauxi group. (3) Overall, many more species are identified when using molecular patterns than when characterizing species in terms of their karyotype. This is particularly obvious for Avahi where chromosome changes only allow to distinguish between an eastern and a western taxon, whereas the molecular data identifies five species within the eastern group and two species within the western one (Andriantompohavana et al. 2007; Zaramody et al. 2006). Similarly, among Propithecus, P. verreauxi/P. coronatus/P. deckeni and P. coquereli share the same karyotype, but the molecular data clearly distinguish P. coquereli as a separate taxon, which constitutes the sister taxon to P. tattersalli. The other three taxa of this group form a distinct clade but further taxonomic differentiation is not possible based on cyt b sequence variation. P. verreauxi, P. coronatus, and P. deckeni as well as morphological hybrid forms between P. coronatus and P. deckeni cluster in the same clade, thus providing no arguments for a classification into distinct species (Pastorini et al. 2001, but see Thalmann et al. 2002 for distinction into subspecies).
Among the eastern Propithecus, P. edwardsi has a specific karyotype, separating it from the other eastern Propithecus taxa, which all share the same karyotype (P. diadema, P. perrieri, P. candidus). The molecular data, however, suggest a possible specific status for P. diadema, P. candidus, and P. perrieri besides P. edwardsi (Mayor et al. 2004).
Our molecular analysis revealed two well-supported mitochondrial clades in P. diadema. One (P. diadema 2) is most likely closer related to P. edwardsi than to its conspecific clade (P. diadema 1). Taking only the genetic distance into account, one would have to split P. diadema into two taxa because the genetic distance is at least as large as for P. tattersalli and P. coquereli. Morphologically, however, no clade-specific differences are known karyotypically; both clades are the same and evidence from nuclear genetic markers is not yet available. Therefore, we think that the current data do not warrant the delineation of a new taxon. Instead, we propose male introgression as a possible cause for the paraphyly of P. diadema (Funk and Omland 2003). We envision three isolated ancestral Propithecus populations along the central and east coast of Madagascar. The southern population refers to proto-P. edwardsi, the central to proto-P. diadema 2, and the northern to proto-P. diadema 1. After a phase of isolation, where population-specific mitochondrial clades evolved, the northern and central populations came into secondary contact. Males of the northern population invaded the central population and reproduced successfully. Hybrid females backcrossed with new invading northern males over generations. As a result, the central population would be swamped with northern nuclear DNA, but would retain their own mitochondria, which was more closely related to that from P. edwardsi than to the mitochondria from the northern population. Today, morphology and nuclear genome of the introgressed central population is very similar or identical to the northern population P. diadema, but its mitochondria are still the original and closely related to P. edwardsi. At one site, Zahamena, individuals of both P. diadema haplogroups are found sympatrically.
Natural hybridization and introgression have recently been recognized as one evolutionary mechanism of speciation for many vertebrate species (Seehausen 2004; Mallet 2005) including primates (review in Arnold and Meyer 2006). However, hybridization between species does not necessarily lead to the formation of new species. In many cases, hybrid offspring suffer from reduced viability or they are sterile. Sex-specific introgressive hybridization is of particular interest because here the result can be the capture of mitochondria or Y chromosomes followed or not followed by nuclear swamping. The genome of the introgressed population or species changes due to horizontal gene transfer without speciation (Zinner et al. 2009a, b).
Indeed, lemurs often hybridize in the wild resulting in horizontal gene transfer (Fausser et al. 2002; Wyner et al. 2002; Pastorini et al. 2009). Therefore, a dichotomic phylogeny might not necessarily indicate a complete interruption of gene flow between different clades, and this becomes particularly relevant when the bootstrap values are very low as it is the case for the topology of Indri. Reticulate evolution was also hypothesized on the chromosomal level, suggesting that in a number of species the chromosomal evolution passed through phases of reticulation, during which chromosome rearrangements could be exchanged among populations of diverging lineages, and cladogenesis, when molecular mutations and/or chromosomal changes have a stronger selective effect (Dutrillaux 1986; Moulin et al. 2008).
We propose a synthetic scheme in which both kinds of events, chromosomal and molecular ones, played a major role during the Indriidae speciation process.
Among Avahi, a chromosomal and a molecular barrier between the eastern and western populations were followed by molecular changes giving rise to numerous species without any chromosomal changes. Among Propithecus, hemiplasic chromosomal changes occurred (Fig. 3) while the molecular evolution split this group into several subgroups or proto-taxa. These ancestral subgroups probably were not yet reproductively isolated and the reproductive barriers appeared later with, or due to, the specific chromosome changes. Proto-P. edwardsi differentiated into a species characterized by chromosomal and molecular changes, while P. diadema experienced only molecular changes which allowed distinguishing at least subspecies: P. d. diadema, P. d. perrieri, and P. d. candidus. Among the P. coronatus, P. verreauxi, P. deckeni/P. coquereli, and P. tattersalli ancestral populations, molecular changes occurred south of the Betsiboka River and gene flow appeared to be maintained between P. verreauxi, P. coronatus, and P. deckeni while north of this river molecular evolution first isolated P. coquereli and P. tattersalli from the P. verreauxi, P. coronatus, and P. deckeni group and later P. coquereli from P. tattersalli. The latter accumulated numerous chromosome changes. We cannot specify precisely if these were at the origin of its divergence from P. coquereli.
Both chromosome and molecular phylogenies are in accordance with the general biogeographical divisions for Madagascar (Martin 2000). The distributions of Propithecus and Avahi follow the traditionally recognized east–west and north–south zoogeographical divisions (Martin 2000; Wilmé et al. 2006; Thalmann 2007; Rumpler et al. 2008). Both mitochondrial and chromosomal phylogenies are consistent with a primary east/west division between Avahi and Propithecus/Indri. Within the eastern region, the Mangoro–Onive River system constituted a major geographical barrier separating the northern from the southern forms. Within the western region, the Betsiboka River also constitutes a major geographical barrier separating northern from southern taxa. The grouping of P. tattersalli and P. coquereli also indicates ancestral transverse relationships between the species living in the eastern and western domain similar to that observed between Lepilemur edwardsi and Lepilemur microdon (Andriaholinirina et al. 2006).
Conclusion
Our chromosomal and molecular analyses of Indriidae are complementary approaches to propose a unified model of the evolutionary history of this lemur family which corresponds to their current zoogeographical distribution. Both approaches show that Avahi diverged in an eastern and western group followed by differentiation on the mitochondrial level but without any chromosomal changes. For the Propithecus/Indri clade, the cladistic chromosomal phylogeny has been retained as the most likely with four hemiplasic rearrangements for which an ancestral polymorphism has to be assumed. Both cytogenetic and molecular phylogenies show a clear distinction between the western (P. verreauxi, P. coquereli, and P. tattersalli) and eastern domain species (P. diadema and P. edwardsi). Neither chromosomal nor molecular data, however, support a separation of the three western Propithecus forms, P. verreauxi, P. coronatus, and P. deckeni, into distinct species. On the other side, both datasets suggest the Mangoro–Onive Rivers as the barrier between P. edwardsi and P. diadema in the eastern Malagasy forests.
Abbreviations
- ALA:
-
Avahi laniger
- AOC:
-
Avahi occidentalis
- CCD:
-
Charge-coupled device
- DAPI:
-
4′,6-diamidino-2-phenylindole
- DMSO:
-
Dimethylsulfoxide
- EFU:
-
Eulemur fulvus
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- h:
-
Heterochromatin
- HSA:
-
Homo sapiens
- IIN:
-
Indri indri
- ML:
-
Maximum likelihood
- MMR:
-
Microcebus murinus
- MP:
-
Maximum parsimony
- mtDNA:
-
Mitochondrial DNA
- NJ:
-
Neighbor joining
- PAUP:
-
Phylogenetic analysis using parsimony
- PCO:
-
Propithecus coronatus
- PCQ:
-
Propithecus coquereli
- PDI:
-
Propithecus diadema
- PED:
-
Propithecus edwardsi
- PTA:
-
Propithecus tattersalli
- PVE:
-
Propithecus verreauxi
References
Andriaholinirina N, Fausser J-L, Roos C et al (2006) Molecular phylogeny and taxonomic revision of the sportive lemurs (Lepilemur, Primates). BMC Evol Biol 6:17
Andriantompohavana R, Lei R, Zaonarivelo JR et al (2007) Molecular phylogeny and taxonomic revision of the woolly lemurs, genus Avahi (Primates: Lemuriformes). Texas Tech Univ Special Publ 51:1–59
Arnold ML, Meyer A (2006) Natural hybridization in primates: one evolutionary mechanism. Zoology 109:261–276
Avise JC, Robinson T (2008) Hemiplasy: a new term in the lexicon of phylogenetics. Syst Biol 57:503–507
Britton-Davidian J, Catalan J, Da Graça RM et al (2005) Chromosomal phylogeny of Robertsonian races of the house mouse on the island of Madeira: testing between alternative mutational processes. Genet Res 86:171–183
Curtis DJ, Velo A, Raheliarisoa EO, Zaramody A, Mueller P (1998) Surveys on Propithecus verreauxi deckeni, a melanistic variant, and P. v. coronatus in north-west Madagascar. Oryx 32:157–164
Delpero M, Masters JC, Cervella P, Crovella S, Ardito G, Rumpler Y (2001) Phylogenetic relationships among the Malagasy lemuriforms (Primates: Strepsirrhini) as indicated by mitochondrial sequence data from the 12S rRNA gene. Zool J Linn Soc 133:83–103
Dobigny G, Ducroz JF, Robinson TJ, Volobouev V (2004) Cytogenetics and cladistics. Syst Biol 53:470–484
Dutrillaux B (1986) Relations chromosomiques entre sous-ordres et infra-ordres, et schéma évolutif général des Primates. In: Evolution chromosomique chez les Primates, les Carnivores et les Rongeurs. Mammalia 50:108–121
Fausser JL, Prosper P, Donati G, Ramanamanjato JB, Rumpler Y (2002) Phylogenetic relationships between Hapalemur species and subspecies based on mitochondrial DNA sequences. BMC Evol Biol 2:4
Funk D, Omland K (2003) Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Ann Rev Ecol Evol Syst 34:397–423
Hill WCO (1953) Primates: comparative anatomy and taxonomy, vol. I. Strepsirrhines. University Press, Edinburgh
Horvath JE, Weisrock DW, Embry SL et al (2008) Development and application of a phylogenomic toolkit: resolving the evolutionary history of Madagascar’s lemurs. Genome Res 18:489–499
Kehrer-Sawatzki H, Cooper DN (2008) Molecular mechanisms of chromosomal rearrangement during primate evolution. Chromosome Res 16:41–56
Lei R, Engberg SE, Andriantompohavana R et al (2008) Nocturnal lemur diversity at Masoala National Park. Texas Tech Univ Special Publ 53:1–41
Mallet J (2005) Hybridization as an invasion of the genome. Trends Ecol Evol 20:229–237
Martin RD (2000) Origins, diversity and relationships of lemurs. Int J Primatol 21:1021–1049
Mayor MI, Sommer JA, Houck ML et al (2004) Specific status of Propithecus spp. Int J Primatol 25:875–900
Mittermeier RA, Louis EE Jr, Richardson M et al (2010) Lemurs of Madagascar, 3rd edn. Conservation International, Arlington
Moulin S, Gerbault-Seureau M, Dutrillaux B, Richard F (2008) Phylogenomics of African guenons. Chromosome Res 16:783–799
Pastorini J, Forstner MRJ, Martin RD (2001) Phylogenetic history of sifakas (Propithecus: Lemuriformes) derived from mtDNA sequences. Am J Primatol 53:1–17
Petter JJ, Albignac R, Rumpler Y (1977) Mammifères lémuriens (Primates prosimiens). In: Faune de Madagascar, vol. 44. CNRS/ORSTOM, Paris, pp 1–512
Posada B, Crandall KA (1998) MODELTEST: testing the model of DNA substitutions. Bioinformatics 14:817–818
Powzyk J, Thalmann U (2003) Indri indri, Indri. In: Goodman SM, Benstead JP (eds) The natural history of Madagascar. University of Chicago Press, Chicago, pp 1342–1345
Qumsiyeh MB, Hamilton MJ, Schlitter DA (1987) Problems in using Robertsonian rearrangements in determining monophyly: examples from the genera Tatera and Gerbillurus. Cytogenet Cell Genet 44:198–208
Robinson TJ, Ruiz-Herrera A, Avise JC (2008) Hemiplasy and homoplasy in the karyotypic phylogenies of mammals. Proc Natl Acad Sci USA 105:14477–14481
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Roos C, Schmitz J, Zischler H (2004) Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci USA 101:10650–10654
Rumpler Y, Albignac R (1979) Cytogenetic of the Indriidae Bennet, 1832, an endemic Malagasy lemur family. In: Chiarelli B, Koen AL, Ardito G (eds) Comparative karyology of primates. Mouton, New York, pp 29–34
Rumpler Y, Dutrillaux B (1990) Chromosomal evolution and speciation in primates. In: RBC, University of the Basque Country (ed) Volume 23, review. Springer, Berlin, pp 1–112
Rumpler Y, Warter S, Ishak B, Dutrillaux B (1988) Chromosomal evolution in Malagasy lemurs: X. Chromosomal banding studies of Propithecus diadema edwardsi and Indri indri and phylogenic relationships between all the species of the Indriidae. Am J Primatol 16:63–71
Rumpler Y, Warter S, Rabarivola C, Petter JJ, Dutrillaux B (1990) Chromosomal evolution in Malagasy lemurs: XII. Chromosomal banding study of Avahi laniger occidentalis (Syn: Lichanotus laniger occidentalis) and cytogenetic data in favour of its classification in a species apart—Avahi occidentalis. Am J Primatol 21:307–316
Rumpler Y, Andriaholinirina N, Warter S, Hauwy M, Rabarivola C (2004) Phylogenetic history of the sifakas (Propithecus: Lemuriformes) derived from cytogenetic studies. Chromosome Res 12:453–463
Rumpler Y, Warter S, Hauwy M, Jean-Luc Fausser JL, Roos C, Zinner D (2008) Comparing chromosomal and mitochondrial phylogenies of sportive lemurs (Genus Lepilemur, Primates). Chromosome Res 16:1075–1250
Schwarz E (1931) A revision of the genera and species of Madagascar Lemuridae. Proc Zool Soc London 1931:399–428
Seehausen O (2004) Hybridization and adaptive radiation. Trends Ecol Evol 19:198–207
Simons EL (1988) A new species of Propithecus (Primates) from Northeast Madagascar. Folia Primatol 50:143–151
Stanyon R, Rocchi M, Capozzi O et al (2008) Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res 16:17–39
Swofford D (2000) PAUP*4.0b.4a. Phylogenetic analysis using parsimony* (and other methods). Sinauer, Sunderland
Tattersall I (1982) The primates of Madagascar. Columbia University Press, New York
Tattersall I (1986) Notes on the distribution and taxonomic status of some subspecies of Propithecus in Madagascar. Folia Primatol 46:51–63
Thalmann U (2007) Biodiversity, phylogeography, biogeography and conservation: lemurs as an example. Folia Primatol 78:420–443
Thalmann U, Geissmann T (2000) Distribution and geographic variation in the western woolly lemur (Avahi ocidentalis) with description of a new species (A. unicolor). Int J Primatol 21:915–941
Thalmann U, Geissmann T (2005) New species of woolly lemur Avahi (Primates: Lemoriformes) in Bemaraha (central Western Madagascar). Am J Primatol 67:371–376
Thalmann U, Kümmerli R, Zaramody A (2002) Why Propithecus verreauxi deckeni and P. v. coronatus are valid taxa—quantitative and qualitative arguments. Lemur News 7:11–16
Vezuli A, Rumpler Y (2000) Lemur chromosomal study with simultaneous R-banding and chromosome painting. Chromosoma 109:214–218
Volobouev VT, AniskinVM LE, Ducroz JF (2002) Patterns of karyotype evolution in complexes of sibling species within three genera of African murid rodents inferred from the comparison of cytogenetic and molecular data. Cytogenet Genome Res 96:261–275
Warter S, Hauwy M, Dutrillaux B, Rumpler Y (2005) Application of molecular cytogenetics for chromosomal evolution of the Lemuriformes (Prosimians). Cytogenet Genome Res 108:197–203
Wilmé L, Goodman SM, Ganzhorn JU (2006) Biogeographic evolution of Madagascar’s microendemic biota. Science 312:1063
Wyner YM, Johnson SE, Stumpf RM, DeSalle R (2002) Genetic assessment of a white-collared x red-fronted lemur hybrid zone at Andringitra, Madagascar. Am J Primatol 67:51–66
Zaramody A, Fausser J, Roos C et al (2006) Molecular phylogeny and taxonomic revision of the eastern woolly lemurs (Avahi laniger). Primate Report 74:9–23
Zinner D, Groeneveld LF, Keller C, Roos C (2009a) Mitochondrial phylogeography of baboons (Papio spp.)—indication for introgressive hybridization? BMC Evol Biol 9:83
Zinner D, Arnold ML, Roos C (2009b) Is the new primate genus Rungwecebus a baboon? PLoS ONE 4:e4859
Zwickl DJ (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. Ph.D. thesis, The University of Texas at Austin
Acknowledgments
We benefited from cooperation with the following persons and entities: the Direction des Eaux et Forêts of Antananarivo and Mahajanga, and the Association Nationale pour la Gestion des Aires Protégées of Antananarivo and Mahajanga (ANGAP) for their permission to capture the animals, to take samples, and for delivery CITES. Thanks to M. Lavaux for her secretarial assistance and Tabitha Price for helping with the English and V. Volobouev for his fruitful help with cladistic phylogeny. Thanks to the Institut Pasteur de Madagascar for providing logistic and technical assistance. The Parc Zoologique de Mulhouse supported the field work of P. deckeni and P. coronatus in order to test the hypothesis of its director, Dr. P. Moisson, that they are able to hybridize. This study was funded by the Association Européenne pour l’Etude et la Conservation des Lémuriens (A.E.E.C.L.) and the DREIF-MENESR (EA3428), and Prof. B. Ludes partly supported the work of Y.R., J.L.F., and N.A.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Fengtang Yang
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Table S1
Samples included in molecular analysis (total 93 samples, sampling sites with geographic coordinates in decimal degree). Seventy-one unique cyt b haplotypes—37 Avahi, 33 Propithecus, and one Indri. *Haplotypes included in phylogenetic tree reconstruction and respective GenBank accession number. (DOC 23 kb)
Table S2
(PDF 42 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rumpler, Y., Hauwy, M., Fausser, JL. et al. Comparing chromosomal and mitochondrial phylogenies of the Indriidae (Primates, Lemuriformes). Chromosome Res 19, 209–224 (2011). https://doi.org/10.1007/s10577-011-9188-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9188-5