Abstract
Drosophila melanogaster HP1-interacting protein (Hip) is a partner of heterochromatin protein 1 (HP1) and is involved in transcriptional epigenetic gene silencing and the formation of heterochromatin. Recently, it has been shown that HP1 interacts with the telomere capping factor HP1/ORC (origin recognition complex)-associated protein (HOAP). Telomeres, complexes of DNA and proteins at the end of linear chromosomes, have been recognized to protect chromosome ends from degradation and fusion events. Both proteins are located at telomeres and prevent telomere fusions. Here, we report the identification and characterization of the Hip-interacting protein Umbrea. We found that Umbrea interacts directly with Hip, HP1 and HOAP in vitro. Umbrea, Hip and HP1 are partners in a protein complex in vivo and completely co-localize in the pericentric heterochromatin and at telomeres. Using a Gal4-induced RNA interference system, we found that after depletion of Umbrea in salivary gland polytene chromosomes, they exhibit multiple telomeric fusions. Taken together, these results suggest that Umbrea cooperates with Hip, HP1 and HOAP and plays a functional role in mediating normal telomere behaviour in Drosophila.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, a large portion of the genome is represented by heterochromatin. Heterochromatin corresponds to relatively gene-poor, late-replicating, repetitive sequences, and heterochromatic regions are predominantly located near centromeres and telomeres. Telomeres, the DNA–protein complexes at the ends of chromosomes, have long been recognized to play important roles in the protection, replication and stabilization of chromosome ends. Chromosome end-capping is an essential function of telomeres. In D. melanogaster, distal chromosome regions (the cap) are capped by a protein complex, which binds the chromosome ends in a DNA sequence-independent manner (Biessmann et al. 1990). In contrast to most eukaryotes, terminal DNA elongation in D. melanogaster does not require telomerase activity, and is mainly provided by the attachment of terminal repeat retrotransposons, He-T-A, TART and TAHRE, to the chromosomal ends (Abad et al. 2004; Biessmann et al. 1992; Sheen and Levis 1994). Proximally are situated subtelomeric telomere-associated sequence (TAS) repeats (Biessmann and Mason 2003). Telomeric proteins play a key role in the protective activity of telomeres and protect chromosome ends from degradation and fusion events (reviewed in Cenci et al. 2005). Telomere dysfunction is now understood to be an important factor in carcinogenesis (Maser and DePinho 2002). Loss of telomere function in human senescent and tumour cells results in the formation of dicentric chromosomes and other abnormalities created through end-to-end fusions (Counter et al. 1992). It is believed that telomeres are fusigenic because they cannot recruit sufficient amounts of telomere capping proteins (van Steensel et al. 1998; de Lange 2002).
In Drosophila, several mutations have been identified that display telomere fusion in larval brain cells (reviewed in Cenci et al. 2005). These include the Su(var)2-5 and caravaggio (cav) genes that encode heterochromatin protein 1 (HP1) and HP1/ORC-associated protein (HOAP), respectively (Fanti et al. 1998; Cenci et al. 2003). In wild type, these proteins protect telomeres from fusion events.
HP1 specificially interacts with HOAP, a DNA-binding polypeptide that plays a role in centric heterochromatin (Badugu et al. 2003). HOAP contains a novel peptide repeat that binds both the chromo shadow domain and hinge domain of HP1 (Badugu et al. 2003). In addition, both HOAP and HP1 interact with the origin recognition complex (ORC; Pak et al. 1997; Badugu et al. 2003). Immunolocalization studies on polytene chromosomes showed that HOAP displays a prominent localization at telomeres but is not enriched in the chromocentre. HOAP co-localizes with HP1 at all telomeres but to a lower extent in discrete regions of the chromocentre (Shareef et al. 2001).
In contrast, HP1, probably the best-studied non-histone chromosomal protein, conserved from yeast to humans (Singh and Georgatos 2002), localizes to centric and telomeric heterochromatin, the banded small fourth chromosome (James et al. 1989) as well as approximately 200 sites throughout euchromatin (James et al. 1989; Sun et al. 2000; Fanti et al. 2003). In Drosophila, HP1 is encoded by the gene Su(var)2-5, a suppressor of position effect variegation (PEV) (Eissenberg et al. 1990) and regulates epigenetic gene silencing and heterochromatin formation by promoting and maintaining chromatin condensation.
The 206-amino-acid (aa) Drosophila HP1 protein contains an N-terminal chromo domain. The chromo domain mediates the association of HP1 and pericentric heterochromatin by binding the N-terminal tail of histone H3 methylated at lysine 9 (H3mLys9) (Bannister et al. 2001; Lachner et al. 2001). The C-terminal chromo shadow domain forms a homodimer (Brasher et al. 2000) and interacts with some chromosomal proteins (Eissenberg and Elgin 2000; Li et al. 2002), including Hip (see below and Schwendemann et al. 2008). The hinge region links the chromo and the chromo shadow domains, is important for nuclear targeting (Smothers and Henikoff 2001), and affects HP1 protein interactions and chromosomal distributions (Badugu et al. 2005).
Recently, we have identified and characterized Hip, an HP1-interacting protein (Schwendemann et al. 2008). Hip and HP1 interact in vitro and in vivo. The HP1 chromo shadow domain is necessary and sufficient for binding of Hip. The interaction is mediated by at least three independent but similar HP1-binding modules of the Hip protein. The presence of three binding surfaces suggests that Hip functions as a bridging protein to cross-connect multiple HP1 proteins, which would contribute to the stabilization of a higher-order chromatin conformation. Hip and HP1 completely co-localize in the pericentric heterochromatin. Both, hip haplo- and triplo-dosage mutations act as dominant suppressors of PEV.
It has been shown that a multiprotein complex is required to establish a heterochromatin structure and that HP1, a key component of these complexes, interacts with a myriad of proteins (reviewed in Li et al. 2002). The results so far suggest that the HP1 protein functions as an adaptor bringing together different proteins in multiprotein complexes. Hence, in order to understand the mechanisms involved, identification of novel proteins that associate with heterochromatin would provide insight into the mechanisms of heterochromatin multiprotein function.
Here, we characterize Umbrea, a Drosophila Hip and HP1-interacting protein. The gene umbrea is identical to the recently described HP6 encoding gene (Greil et al. 2007). In this study, HP6 and three other proteins (HP3, HP4 and HP5) were characterized as four novel Drosophila heterochromatin proteins. Greil et al. used the Drosophila Interaction Database (Giot et al. 2003) to search for proteins that directly interact with HP1 and named these proteins HP3, HP4 (identical to Hip), and HP5. However, on the basis of information in the Drosophila Interaction Database, Greil et al. propose that the fourth protein, HP6 (identical to Umbrea described here), does not directly bind HP1 but binds HP3, HP4, HP5 and HOAP. Although Greil et al. name the novel heterochromatin protein HP6, we refer to the official FlyBase name umbrea for the gene that we describe and characterize in this report.
Umbrea contains only a chromo shadow domain. Hip, HP1 and Umbrea can be co-immunoprecipitated from nuclear extracts. Umbrea directly binds to HP1 and Hip in vitro. The interaction between HP1 and Umbrea is mediated by their chromo shadow domains. Umbrea, like HP1, binds the same three HP1-binding modules of the Hip protein. Umbrea co-localizes with HP1 and Hip in the pericentric heterochromatin and at telomeres and HP1 controls genomic targeting of Umbrea. Mutations in umbrea cause telomeric associations of polytene chromosomes. This finding suggests a function of Umbrea in telomere protection in wild type. The mechanisms of telomere protection in Drosophila are not well understood. The characterization of Umbrea will contribute to the understanding of Drosophila telomeres.
Materials and methods
Cloning of umbrea
Fragment umbrea (coding for Umbrea protein) was amplified by PCR using total genomic DNA from adult D. melanogaster and primers umbrea-fwdBamHI (gcggcggatccatATGCCCAGCTCCACTTT) and umbrea-revXhoI (cgcggcctcgagGGCATTTCGTGATCGTTT). The primers contained sites for BamHI or XhoI and the PCR fragment was cloned into pET21c (Novagen/EMD Chemicals, San Diego, CA, USA).
Antibodies and chromosome immunostaining
Antisera anti-Umbrea I and III were produced against the protein encoded by the umbrea fragment described above. The His-tagged Umbrea recombinant protein was expressed in BL21(DE3) Escherichia coli cells using construct pET21c-umbrea. The protein was purified using Ni-NTA columns (Novagen) and standard procedures. The eluate fraction was used as an immunogen in two guineapigs by Pineda Antikoerper Service (Berlin, Germany). The specificity of the anti-Umbrea antibodies was tested by immunoblots using Drosophila salivary gland nuclear extract (for preparation see Lehmann and Korge 1995). Antibodies from these sera were used in chromosome staining and immunoprecipitation reactions.
Whole-mount immunostaining of salivary glands was carried out essentially as described in Siegmund and Korge (2001). Polytene chromosomes were stained as described in Lehmann and Korge (1996) using monoclonal anti-HP1 antibody (a gift of S. C. R. Elgin, Washington University, St. Louis, MO, USA.), anti-Umbrea I antiserum described above, and anti-Hip II antiserum (Schwendemann et al. 2008). Cy3-conjugated AffiniPure goat anti-guinea pig (Jackson ImmunoResearch, West Grove, PA, USA), Alexafluor 488 goat anti-rabbit IgG conjugate (Molecular Probes/Invitrogen, San Diego, CA, USA), and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch) were used as secondary antibodies. DNA was counterstained with the dye Hoechst33258. Preparations were examined with a Zeiss Axiophot fluorescence microscope and a Quantix (Photometrics) video camera. Images were processed with Photoshop 7.0.
Immunoprecipitation and direct protein interaction assays
Co-immunoprecipitation reactions containing 90 μl of salivary gland nuclear extract (preparation described in Lehmann and Korge 1995) and 50 μl each of anti-Umbrea antisera I and II were performed essentially as described in Schwendemann et al. (2008), and SDS-PAGE and immunoblotting were performed using standard procedures. Co-immunoprecipitation of HP1 and Hip was detected using anti-HP1 and anti-Hip antibodies. As a negative control, a combination of pre immune sera I and II was used in the immunoprecipitation reaction.
GST-fusion proteins of full-length (GST-Hip) and truncated forms of Hip used for different GST pull-down tests are shown in Fig. 1d and described in Schwendemann et al. (2008). The GST-HP1 fusion construct was made by cloning into pGEX2T the sequence that corresponds to HP1 amino acids 1 to 216. Umbrea constructs were made by cloning fragments that correspond to amino acids 1–19 (for GST-Umbrea-N), 20–74 (for GST-Umbrea-cs), and 75–106 (for GST-Umbrea-C) into pGEX3X. Full-length Umbrea (amino acids 1–106) was subcloned in pGEX2T. Generation of His-tagged HP1 fragments (HP1-cd-His, HP1-cs-His, HP1-hinge-His) is described in Schwendemann et al. (2008). For GST pull-down assays, His6-tagged recombinant proteins were expressed in BL21(DE3) and crude bacterial cell extracts loaded on glutathione Sepharose-4B beads (Amersham Pharmacia/GE Healthcare, Piscataway, NJ, USA) immobilized GST-fusion proteins. GST pull-down assays were performed according to standard protocols. Further details can be obtained upon request. Bound proteins were eluted and fractionated by SDS-PAGE, and the presence of His-tagged proteins was assayed by immunoblotting using anti-His tag specific antibody (Novagen).
Umbrea binds directly to Hip and HP1 in vitro and the three proteins are associated with one another in a protein complex in vivo. In addition, Umbrea is a protein interaction partner of HOAP. Umbrea uses the same binding modules in Hip as HP1 and HP1 interacts with Umbrea using its chromo shadow domain. Full-length Umbrea, Hip, HP1 and HOAP proteins (depicted in; a were expressed in bacteria and assayed for their ability to interact with each other in; b Note: Hip contains three HP1-binding interfaces (I, II and III; described in Fig. 3), whereas HOAP contains a HMG-like domain and three copies of a proline-containing repeat (RP1-3) (Shareef et al. 2001); b A GST-Hip, GST-HP1 and GST-HOAP fusion protein, or GST alone, were used in a GST pull-down assay and analysed for their ability to retain recombinantly expressed His-tagged full-length Umbrea. Eluted proteins were probed with an anti-His tag antibody. Umbrea interacts with Hip, HP1 and HOAP; c Anti-HP1 and anti-Hip western blot analysis after immunoprecipitation (IP). Nuclear extract from larval salivary glands was immunoprecipitated using a combination of both anti-Umbrea antibodies, or mock-precipitated using the corresponding preimmune sera. Mock precipitation with preimmune sera did not retain Hip and HP1, respectively, indicating that Umbrea interacts specificially with Hip and HP1 in vivo; d The Hip protein sequence contains numerous charged amino acid residues, such as K, R, E and D (bold type; positively and negatively charged amino acids are underlined and in italics, respectively). (Figure modified after Fig. 4 in Schwendemann et al. 2008.) As described in Schwendemann et al. (2008), Hip contains three HP1-binding interfaces depicted below (I, II and III). The Hip N- and C-terminal fragments used for the following experiments are indicated.; e As in (B), His-tagged full-length Umbrea was used as an input in GST pull-down assays with different Hip N-terminal (GST-N-Hip, GST-N1-Hip, GST-N2-Hip, GST-N3-Hip and GST-N4-Hip) or C-terminal (GST-C-Hip, GST-C1-Hip, and GST-C2-Hip) fragments. Western blot analysis with anti-His tag antibody revealed that fragments N-Hip, C-Hip, N1-Hip, N2-Hip and N4-Hip, but not N3-Hip, C1-Hip or C2-Hip, interact with Umbrea. Three sequences that are necessary for Umbrea binding are underlined; e. Note: these sequences are identical to the three HP1-binding interfaces of Hip (Schwendemann et al. 2008); f To characterize the interaction between Hip and Umbrea, His-tagged full-length Hip was used as an input in GST pull-down assays but Umbrea N-terminal (GST-N-Umbrea), C-terminal fragments (GST-C-Umbrea), or the chromo shadow domain (GST-cs-Umbrea) were immobilized on glutathione agarose beads. The Umbrea chromo shadow domain but not N- or C-terminal regions is sufficient to interact with Hip; g Same conditions as before, but this time full-length Umbrea was fused to GST (GST-Umbrea) and GST pull-down assays with truncated forms of His-tagged HP1 (HP1-cd-His, HP1-hinge-His or HP1-cs-His) were performed. The HP1 chromo shadow domain (HP1-cs) but not the HP1 chromo domain (HP1-cd) or the hinge region (HP1-hinge) is necessary to interact with Umbrea. Finally, as described above, but only the chromo shadow domain of Umbrea was fused to GST (GST-cs-Umbrea) and analysed for the ability to pull down the HP1 chromo shadow domain (HP1-cs-His). Taken together, the chromo shadow domains of both Umbrea and HP1 are sufficient to mediate a direct interaction between the two proteins
To generate a GST-HOAP fusion construct the cav coding region was amplified with the primers cav-fwdBamHI (gccgcggatccATGTCGGGGACGCAAAT) and cav-revBamHI (cggccggatccTCAGGCTATTGAGGTAG) using the cav cDNA (clone LD09927 obtained from BDGP) as template. Primers contain sites for BamHI (as indicated; lowercase letters) and, after digestion with BamHI, the PCR fragment was cloned into pGEX2T (Amersham Pharmacia).
Drosophila strains
To examine the binding of Umbrea in the absence of HP1 we used the HP1 null mutant Df(2L)TE128–22/Su(var)2-5 04. These trans-heterozygous larvae were generated by crossing Df(2L)TE128–22/CyOGFP and Su(var)2-5 04 /CyOGFP obtained from G. Reuter (Universität Halle, Germany). Generation of strain hip 41 carrying a deletion in the hip gene is described in Schwendemann et al. (2008). Umbrea mutant line P(GT1)CG15636 BG01429 (w 1118 ; net1 P(GT1)CG15636 BG01429 dp BG01429 /In(2LR)Gla, wg Gla-1 Bc 1) is a P-element insertion line and was obtained from the Bloomington Drosophila Stock Center. The Umbrea encoding gene CG15363 consists of a single exon located in an intron of the larger dumpy (dp) gene. We cannot rule out that the expression of dp is affected in this mutant. Any transcript that could arise from P(GT1)CG15636 BG01429 is unlikely to produce a functional Umbrea protein, because the open reading frame starting at the next methionine would have a truncated chromo shadow domain. The chromo shadow domain is the only recognizable domain in Umbrea.
For stage- or tissue-specific umbrea RNAi knock-down we used the transformant line 13072 from the Vienna Drosophila RNAi Center (Dietzl et al. 2007) which carries the UAS-umbrea RNAi construct (construct ID: 4434) inserted on the third chromosome. We crossed males carrying this umbrea RNAi construct with different Gal4 driver strains. We used act-Gal4 (act-Gal4/TM6B) and da-Gal4 (da-Gal4/TM6B) Gal 4 driver lines for ubiquitous RNAi knock-down. To target gene interference to salivary glands we used homozygous G61-Gal4 (G61/G61). The enhancer trap line G61-Gal4 was used as an embryonic salivary gland driver. Expression of Gal4 in this line starts at embryonic stage 16 (U. Hinz, personal communication 2001).
Results
Identification of Umbrea, a Hip- and HP1-interacting protein
In an effort to identify genes that are regulated by the transcription factor JUMU we assessed global changes in gene transcription following overexpression of jumu. Also known as Domina (Dom), jumu encodes a Drosophila winged-helix domain transcription factor of pleiotropic function. It is involved in developmental processes such as neurogenesis, in development of eyes and wings, and in general features such as vitality and fertility. It modifies chromatin structure and the gene jumu is a haplo-suppressor/triplo-enhancer of PEV (Cheah et al. 2000; Strodicke et al. 2000). In the microarray experiment the highest magnitude in expression level changes was observed for hip. Information from the Drosophila interaction database (Giot et al. 2003) suggested that Hip, the protein product of hip, interacts with three other protein partners. The interaction of one partner of Hip (HP1-interacting protein), the heterochromatin protein 1 (HP1), was characterized recently. Hip is a protein that interacts with HP1 and both proteins completely co-localize in the pericentric heterochromatin. Hip cooperates with HP1 in chromatin remodelling and gene silencing (Schwendemann et al. 2008). To gain more functional information about Hip we characterized another potential interacting partner of Hip that is encoded by the gene umbrea.
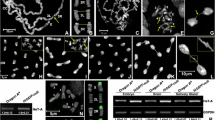
For initial experiments we amplified the coding region of umbrea by PCR according to predicted sequence of the annotated umbrea transcript. As a template we used total genomic DNA since the Umbrea-encoding gene consists of a single exon. We verified the putative Hip/Umbrea interaction by in vitro GST pull-down assays. A GST-Hip fusion protein (Schwendemann et al. 2008) was expressed in bacteria and we then tested whether this polypeptide is able to co-precipitate recombinant Umbrea protein contained in a complex E. coli cell extract. For ease of detection we expressed an N-terminal histidine (his)-tagged form of Umbrea (Umbrea-His). We then analysed the eluate of the GST pull-down in immunoblots using anti-His tag specific antibody. As shown (Fig. 1b, upper image), GST-Hip interacts very efficiently with Umbrea. As a negative control, GST alone shows no interaction with Umbrea. Together, our initial interaction experiments confirm that the protein Umbrea is indeed capable of binding Hip.
Next, we asked whether Umbrea is able to interact with HP1, since the chromo shadow domain is the only recognizable domain in Umbrea (see below). This chromo shadow domain is very similar to the HP1 chromo shadow domain (see below and alignment in Fig. 2b). The HP1 C-terminal chromo shadow domain is known to mediate a HP1 homodimer (Brasher et al. 2000). We therefore speculated that Umbrea might be able to heterodimerize with HP1, despite the fact that this interaction is not supported by information from the Drosophila interaction database (Giot et al. 2003). To test this, we repeated the GST pull-down test described above using immobilized GST-HP1 and recombinant Umbrea-His. As shown (Fig. 1b, lower image), Umbrea is indeed able to bind HP1.
The structure of the umbrea gene and the Umbrea protein; a (Upper) A portion of the genomic region containing the umbrea gene located in the first intron of the >200 kb dumpy gene on chromosome 2L is shown. The genomic position is indicated. Transcription start sites (arrows), the insertion site of the P(GT1)CG15636 BG01429, and the predicted umbrea and dumpy transcripts are indicated. Predicted exon/intron structure is marked with boxed exons. (Lower) Enlarged view of the umbrea transcription unit. Note, umbrea transcript consists of a single exon and encodes a 106-amino-acid protein; b Sequence comparison of Drosophila melanogaster Umbrea, HP1 chromo shadow domain (HP1-cs; amino acids 143 to 206), and HP1 chromo domain (HP1-cd; amino acids 20 to 77). Amino acid residues that are similar are shaded grey. Identical amino acids are in bold. The chromo shadow domain is the only recognizable domain in Umbrea
One possibility to explain these protein–protein interactions is that Umbrea and Hip as well as Umbrea and HP1 are associated in a protein complex that contains both (or even all three) proteins in vivo. To confirm in vivo the interaction between Umbrea, Hip and HP1, we therefore prepared protein extracts from salivary gland nuclei and performed immunoprecipitation (IP) assays with a combination of the Umbrea I and II antibodies (described below) and then analysed the IP fraction by immunoblot with Hip- and HP1-specific antibodies. As can be seen in the immunoblot in Fig. 1c, the Umbrea antibody efficiently co-immunoprecipitated Hip and HP1 from these extracts. To test whether this effect was specific, we repeated the immunoprecipitation experiment under same conditions, omitting the Umbrea antibody and using the corresponding preimmune serum instead. However, Hip and HP1 were not found to be precipitated by the preimmune serum (Fig. 1c). Taken together, these results strongly indicate that the interactions between Umbrea and Hip as well as between Umbrea and HP1 are specific and that the proteins are associated in a protein complex in vivo. However, at this point we cannot rule out the possibility that the co-precipitation of HP1 is rather due to an indirect interaction via Hip than a direct binding of Umbrea/HP1.
As described above, four Drosophila heterochromatin proteins have recently been identified, of which three (HP3, HP4 (identical to Hip), and HP5) were shown to directly bind HP1 (Greil et al. 2007). In contrast with our results, Greil et al. propose that the fourth protein (HP6, identical to Umbrea described here) may not directly bind HP1. Here, we clearly show that Umbrea is a direct interaction partner of HP1.
Interestingly, according to the Drosophila interaction database (Giot et al. 2003), the umbrea-encoded protein is predicted also to interact with HOAP, a protein that itself binds to HP1 (see Introduction). To determine whether HOAP is capable of directly interacting with Umbrea, we carried out a GST pull-down experiment with bacterially expressed GST-HOAP fusion protein to test the co-precipitation of Umbrea-His. As HP1 interacts directly with HOAP (Badugu et al. 2003), we used HP1-His as a positive control. In our experiments, Umbrea (Fig. 1b, lower image) and HP1 (not shown) co-precipitated with HOAP. As a negative control, GST alone shows no interaction with Umbrea and HP1. Altogether, these results establish that Umbrea has at least three protein interacting partners: Hip, HP1, and HOAP.
Analysis of the umbrea gene
According to the Berkeley Drosophila Genome Project (BDGP) annotation of the Drosophila genome, the gene umbrea consists of a single exon and is located in the >13.5 kb first intron of the >200 kb dumpy (dp) gene (see Fig. 2a). The BDGP annotation predicts one umbrea transcript of 321 nt. This is further supported by a BDGP full insert cDNA sequence (IP05759). The conceptual translation results in a 106-aa protein. Interestingly, Umbrea consists largely of a chromo shadow domain. We compared the amino acid sequence of Umbrea with the chromo-(HP1-cd) and chromo shadow domain (HP1-cs) of HP1 (Fig. 2b). Clearly, the only recognizable domain in Umbrea is a chromo shadow domain, and sequence comparison leads to 50.8% identity and 75.4% similarity between the HP1 and Umbrea chromo shadow domains. In the remaining amino acids (18 amino acids N-terminal and 23 amino acids C-terminal of the chromo shadow domain) we find no homology to any other known protein (not shown). Hiatt et al. (2007) propose that the gene umbrea is a result of a duplication of the HP1 encoding gene and that umbrea has undergone neofunctionalization, thereby acquiring a novel function.
To analyse the expression of the Umbrea protein, we immunized guineapigs with bacterially expressed GST-Umbrea fusion protein. Both of the two independent antisera obtained detect a protein of ∼15 kDa on immunoblots of Drosophila salivary gland nuclear protein extracts (not shown). The protein size corresponds well with the predicted size for Umbrea of 11.8 kDa. These findings confirm that umbrea is expressed and translated.
Characterization of Umbrea/Hip interaction
Recently, we have identified three HP1-binding modules within the 106-aa sequence of Hip. Each of these sequences appear to be sufficient for HP1 binding (Schwendemann et al. 2008). To characterize the Umbrea/Hip interaction more fully, we next characterized the Umbrea binding sequences within Hip and asked whether Umbrea uses the same three interacting modules of Hip for binding as HP1. To test this, we used several truncated forms of Hip fused to GST (GST-N-Hip, -N1-Hip, -N2-Hip, -N3-Hip, N4-Hip, C-Hip, C1-Hip, and C2-Hip; depicted in Fig. 1d) and performed GST pull-down assays with Umbrea-His as input. Four constructs derived from the N-terminal half of Hip (N-, N1-, N2-, and N4-Hip), showed binding to Umbrea-His. A fifth N-terminal construct, N3-Hip, lacked the ability to bind Umbrea. From the three C-terminal fragments tested (GST-C-Hip, -C1-Hip, C2-Hip) only the longest peptide (C-Hip) showed binding to Umbrea-His (Fig. 1e). Together, this binding behaviour is in perfect agreement with the binding of HP1 to Hip. Both proteins, HP1 and Umbrea, use the same three binding modules within the Hip sequence. Each of these sequences appears to be sufficient for Umbrea binding. This cooperative binding behaviour serves to cross-link multiple HP1 and Umbrea molecules. Such a mechanism could contribute to the stabilization and maintenance of heterochromatin.
We next used the same GST pull-down technique to map the minimal sequence in Umbrea responsible for interaction with Hip. To address this, we expressed several subdomains of the Umbrea coding region (depicted in Fig. 1a) fused to GST and used His-tagged Hip as input. The results (Fig. 1f) show that the binding activity is located in the middle of the Umbrea protein (amino acids 20 to 83; GST-cs-Umbrea) containing the shadow domain. The first 20 amino acids of Umbrea (GST-N-Umbrea) and C-terminal part (amino acids 83 to 106; GST-C-Umbrea) are not essential for this interaction. Altogether, these results indicate that the 63 amino acids of the Umbrea chromo shadow domain are necessary and sufficient for binding Hip.
Characterization of Umbrea/HP1 interaction
The interaction of Umbrea with HP1 was further characterized in subsequent GST pull-down assays. We expressed Umbrea fused to GST and several his-tagged subdomains of the HP1 protein (Fig. 1a) to map the domain in HP1 responsible for interaction with Umbrea. These experiments revealed that the C-terminal chromo shadow domain of HP1 is sufficient for Umbrea/HP1 interaction (HP1-cs-His) (Fig. 1g). In contrast, the N-terminal 86 amino acids of HP1 that contain the chromo domain (HP1-cd-His) and the hinge region (HP1-hinge-His) do not show interaction with Umbrea, indicating that the HP1 chromo shadow domain is necessary and sufficient for binding Umbrea. This result is underlined by our final experiment. We minimally reduced both proteins to the chromo shadow domain and repeated the GST pull-down test with GST-cs-Umbrea and HP1-cs-His (Fig. 1g, right). Taken together, the Umbrea and HP1 chromo shadow domains are sufficient for the direct HP1/Umbrea protein–protein interaction.
Umbrea is co-distributed with HP1 and hip
On the basis of our protein interaction studies with Hip and HP1 we expected that Umbrea is also a heterochromatin-associated protein. In order to test this, we immunostained salivary gland polytene chromosomes with anti-Umbrea antibody. To identify chromosomal loci that are targets of Umbrea, we counterstained the chromosomes with the DNA dye Hoechst 33258. Umbrea is enriched in pericentric heterochromatin and concentrated in the chromocentre and the mostly heterochromatic small fourth chromosome and shows staining at telomeres (Fig. 3a). Strikingly, the staining is very strong at telomeres of the left and right arms of the second chromosome and of the right arm of the third chromosome. In addition, Umbrea localizes to hundreds of sites along the arms of the chromosomes. Interestingly, Umbrea shows binding to the nucleolus.
Immunolocalization of Umbrea, Hip and HP1 on polytene chromosomes; a Chromosomes from late third-instar larvae salivary glands were immunostained with anti-Umbrea antibody (red) and counterstained with the DNA dye Hoechst 33258 (green). Umbrea is localized in the heterochromatic chromocentre (boxed, see magnification), at telomeres (arrowheads), and at the mostly heterochromatic fourth chromosome. Umbrea shows a uniform distribution and binds to hundreds of loci along the arms of polytene chromosomes. In addition, Umbrea is enriched in the nucleolus (asterisk); b Umbrea, Hip and HP1 co-localize in the heterochromatic chromocentre and at telomeres. Chromosomes were immunostained with anti-Umbrea antibody (red) and anti-Hip antibody (green) or anti-HP1 antibody (green). Sites where Umbrea and Hip (or HP1) co-localize appear yellow in the merged image. The yellow signals clearly show that both proteins, Umbrea and Hip, co-localize at telomeres, at the chromocentre and in the nucleolus (asterisk). However, separate green and red signals show that several Umbrea and Hip (or HP1) sites are not identical
As described in detail in the introduction, HP6 (identical to Umbrea), was recently not found to bind directly to HP1 (Greil et al. 2007). For localization studies, Greil et al. used Drosophila Kc cells transfected with DamMyc-tagged HP6 to show the localization of HP6. Whereas other heterochromatin proteins are clearly enriched in the chromocentre, HP6 showed overall nuclear staining. In part, this result is in good agreement with our polytene chromosome staining: we also find that Umbrea binds to many loci along the chromosomal arms. However, in our experiments, Umbrea clearly shows an enrichment to the pericentric chromocentre.
Because various lines of evidence suggest that Umbrea, Hip and HP1 act as partners, we next asked whether the direct interaction of the proteins shown above might reflect a chromosomal co-distribution. We expected the proteins to co-localize in a chromatin-associated complex. To identify chromosomal loci that are targets of Umbrea and Hip, we doubly immunostained polytene chromosomes with anti-Umbrea and anti-Hip antibodies (Fig. 3b). Both proteins completely co-localize in the chromocentre, at telomeres, and on the fourth chromosome. Both Umbrea and Hip show binding to the nucleolus. In contrast, along the arms of polytene chromosomes, several binding sites of Umbrea and Hip are not identical. Given the co-localization of Umbrea and HP1 in the heterochromatic chromocentre, at the telomeres, and at many sites along the arms of all polytene chromosomes (Fig. 3b), we conclude that Umbrea, Hip and HP1 interact not only in vitro but also in vivo. Our finding confirms the results of the DamID large-scale mapping technique in transfected cell culture Drosophila Kc cells performed by Greil et al. (2007). In their experiment, they find that the chromosomal distribution of Umbrea and Hip, among other heterochromatin proteins, is very similar to that of HP1. For each of these proteins, they show conspicuous binding in pericentric regions of the major chromosomes and on the heterochromatic chromosome 4.
On the basis of our protein interaction studies we next asked whether the binding of Umbrea to chromatin depends on the localization of its interacting partners Hip and HP1. To test this, we used HP1-deficient third-instar larvae (Fanti et al. 1998; Lu et al. 2000) and the hip mutant line hip 41 (Schwendemann et al. 2008). Binding of Umbrea to chromatin is dramatically reduced in HP1-deficient larvae (Fig. 4a) in comparison with control stainings of chromosomes from wild-type larvae. This result suggests that binding of Umbrea to heterochromatin and to specific sites within the euchromatin depends on HP1 function. In contrast, the chromosomal distribution of Umbrea along the arms of polytene chromosomes appears to be unaffected in hip 41 homozygous larvae. Interestingly, in this mutant, localization of Umbrea to the chromocentre was reproducibly weaker than in wild-type control larvae, suggesting that binding of Umbrea to the chromocentre depends on Hip (Fig. 4a). We conclude that HP1 is required for chromosomal targeting of Umbrea, whereas Hip—apart from the heterochromatic chromocentre—appears not to be essential for Umbrea chromosomal association.
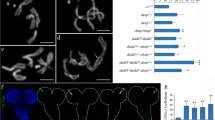
a Binding of Umbrea depends on HP1 but not on Hip. Polytene chromosomes are stained with anti-Umbrea antibody and counterstained with Hoechst. Umbrea binding is strongly reduced in Su(var)2-5-deficient larvae. In these larvae almost no HP1 staining could be detected (not shown). In hip 41 homozygous larvae virtually no Hip protein could be detected (shown in Schwendemann et al. 2008). In these larvae the binding of Umbrea appears unaffected along the arms of the chromosomes but is reduced at the chromocentre; b On chromosomes, chromatin-associated Umbrea is significantly reduced after salivary gland-specific RNAi depletion of Umbrea. The umbrea RNAi line 13072 under the control of UAS and the salivary gland specific driver line G61-Gal4 were used. Polytene chromosomes from umbrea RNAi mutants are stained with anti-Umbrea antibody (red) and counterstained with the DNA dye Hoechst 33258 (green); c Umbrea protein levels are reduced after Umbrea RNAi knock-down. Immunoblots of salivary gland protein extracts of control and salivary gland umbrea RNAi animals were probed with anti-Umbrea antiserum. As a loading control, we used anti-β-actin-antibody (abcam). In RNAi mutants Umbrea protein levels are dramatically reduced
Mutations in Umbrea cause telomeric fusions
To analyse the umbrea function we used the P-element mutant line P(GT1)CG15636 BG01429 from the Bloomington Stock Center. The P-element is inserted into the open reading frame, truncating the predicted protein (see Fig. 2a). Greil et al. (2007) used this line to test a possible role of the corresponding protein as a suppressor of PEV. They were unable to detect a significant effect in this mutant, suggesting that it is not needed for heterochromatic gene silencing. The umbrea P-element mutant line is homozygous lethal. In a lethality test, we find that mutants homozygous for the P-element insertion die during embryogenesis. However, a few larvae reach the first larval stage and die (not shown). It should be noted that the gene coding for Umbrea resides in an intron of the dumpy locus (see Fig. 2a). It is therefore possible that the lethality of the P-element mutant is due to changes in the expression of dumpy.
Given the protein–protein interaction of Umbrea with HP1 and HOAP described above, we speculated that Umbrea has a common functional mechanistic basis. Mutations of the Su(var)2-5 and cav genes that encode HP1 and HOAP, respectively, cause extensive telomere–telomere fusions in larval brain cells, indicating that HOAP and HP1 are required for telomere capping (Fanti et al. 1998; Cenci et al. 2003). But the lethality of the umbrea P-element mutant at embryonic stages precludes a systematic functional analysis during development and does not allow cytological analysis of mitotic chromosomes of larval brain cells. To circumvent this limitation, we studied the role of Umbrea using a Gal4-inducible RNA interference (RNAi) system, which allows for depletion of Umbrea in a tissue-specific and developmental stage-specific manner. We used the umbrea-specific RNAi line 13072 (Dietzl et al. 2007) under the control of UAS. To deplete Umbrea, we crossed the line with an act-Gal4/TM6B and a da-Gal4/TM6B line, respectively. These driver lines express Gal4 ubiquitously during development. Animals bearing both umbrea RNAi construct and Gal4 driver construct chromosomes can easily be recognized because they do not carry the dominant larval marker Tubby (Tb) present on TM6B. All obtained larvae contained the Tubby marker but not the Gal4 driver chromosome, indicating that the lethality after RNAi depletion of Umbrea occurred at the embryonic stage. This result confirms the mutant phenotype of the umbrea P-element mutant line.
Next, we examined the effect of Umbrea depletion in salivary gland tissue in third-instar larvae. We crossed 13072 with the enhancer trap line G61-Gal4. This line was used as an embryonic salivary gland driver. Expression of Gal4 in this line starts at embryonic stage 16, when cell division in salivary glands is completed (U. Hinz, personal communication 2001). We verified umbrea RNAi knock-down in resulting larval progeny by immunostaining salivary glands with anti-Umbrea antibody. In wild-type, Umbrea shows a nuclear staining with typically bright speckles. Compared with those in control animals, after Umbrea RNAi depletion salivary glands appear smaller in size (Fig. 5a, left image) and nuclear Umbrea staining is strongly reduced (Fig. 5a, right image). Consistent with this, Umbrea protein level is dramatically reduced in immunoblots with salivary gland nuclear extracts and Umbrea-specific antibody (Fig. 4c). By immunohistochemistry, we find that Umbrea staining at polytene chromosomes is significantly reduced after Umbrea RNAi knock-down, confirming the umbrea RNAi mutant condition (Fig. 4b). Interestingly, the chromosomal distributions of both HP1 and Hip appear to be unaffected and comparable to wild-type (not shown). We analysed polytene chromosomes of G61/+;13072/+ third instar larvae and, interestingly, after RNAi depletion of Umbrea, we found frequent telomere–telomere attachments. In 43.5% of the nuclei tested we find telomere association events (Table 1 and Fig. 5b). To our surprise, the corresponding negative control line G61/G61 also displayed telomeric attachments and the frequencies (20%) were not comparable to KochiR wild-type controls where the frequencies of telomeric associations were very low (3%). We speculate that in the homozygous driver line G61/G61 chromosome ends stick together in an abnormal way owing to a tremendous overexpression of Gal4 protein. Nevertheless, the feature of telomere attachments is significantly higher in progeny carrying both the Gal4 driver and the umbrea RNAi construct. The frequency of multiple attachments is especially enhanced involving more than two telomeres (see Table 1 for detailed information). To rule out the possibility that elevated effects of telomere fusions might be due to unspecific interactions of Gal4 with telomeres now depleted in Umbrea, we stained these polytene chromosomes with anti-Gal4 specific antibody. With these antibodies we find no staining, suggesting that overexpression of Gal4 has no influence on chromosome structure (not shown).
Depletion of Umbrea causes attachment of polytene chromosome telomeres; a Reduction in Umbrea levels in larval salivary glands by RNAi. (Left images) Salivary glands of mutant salivary gland umbrea RNAi (G61-Gal4/+; 13072/+) third-instar larvae appear smaller in size than in control (G61-Gal4/+; +/+). (Right images) Expression of Umbrea in whole larval salivary glands of control animals and after Umbrea RNAi depletion as indicated by antibody staining against Umbrea. Note the strong nuclear staining in wild type with bright speckles (the boxed areas are shown as enlargement of two nuclei); b Depletion of Umbrea causes polytene chromosome telomere–telomere associations. Fluorescent Hoechst 33258-stained polytene chromosomes after reduction of Umbrea levels by UAS-RNAi-umbrea and the salivary gland-specific driver G61. Sites of attached telomeres are marked (arrow). Boxed areas are shown in enlarged view (right) of the telomere fusion. A polytene chromosome nucleus displays triple telomere attachment (upper panel). Below, all telomeres are fused (arrow), forming a structure reminiscent of a chromocentre (cc arrow)
On closer analysis of the telomere fusion events of polytene chromosomes, our data indicate that all the chromosomes can participate in telomere–telomere associations, albeit with different frequencies. The telomeres of the X chromosome and of the left arm of the third chromosome (3L) appear to participate in telomere attachments more often (not shown). This behaviour is in good agreement with the Umbrea telomere association described above. Umbrea is localized at the telomeres of all chromosomes. But given the predominant Umbrea localization at the 2R, 2L and 3R chromosomes described above, we speculate that the RNAi-induced mutant phenotype is more pronounced at the 3L and X chromosomes as a consequence of the Umbrea depletion.
In contrast to polytene chromosomes, no telomere fusions were detected in mitotic chromosomes after examination of colchicine-treated and non-colchicine-treated brain squashes from larvae bearing the umbrea RNAi mutant. We tested different neuronal specific Gal4 driver lines, but in no case were mitotic abnormalities detected.
However, given the telomere–telomere fusion of polytene chromosomes after RNAi depletion of Umbrea and the protein–protein interaction of Umbrea with HOAP and HP1, we argue that Umbrea is required for telomere capping and to prevent telomere end-to-end fusion in Drosophila.
Discussion
Our study identified and characterized the heterochromatin protein Umbrea by searching a yeast two-hybrid database for predicted interacting partners of the previously characterized HP1-interacting protein Hip (Schwendemann et al. 2008). We not only confirmed the predicted interaction of Umbrea and Hip but we also found that Umbrea is able to interact with HP1. This direct interaction is not reported from the Drosophila interaction database (Giot et al. 2003).
In contrast to our results, Greil et al. performed no additional protein–protein interaction studies to verify the predicted interactions. For localization studies, Greil et al. used epitope-tagged HP6 and HP1 in transfected Drosophila Kc cells. Whereas HP1 is enriched at the heterochromatic chromocentre, for HP6 localization they found a uniform nuclear staining. However, they did not detect a clear co-localization of HP6 and HP1 in the chromocentre. In a different experiment, Greil et al. used the DamID large-scale mapping technique in transfected cell culture Drosophila Kc cells for co-localization studies with HP1 (Greil et al. 2007). In contrast, in this experiment they found binding of HP6 in pericentric regions of the major chromosomes and on the small chromosome 4. HP6 localization was only subtly affected after HP1 depletion. On the basis of this result, Greil et al. speculate that an additional interaction might play a key role in targeting HP6 to heterochromatin. To functionally characterize HP6, Greil et al. tested whether mutation of HP6 is a suppressor of PEV. However, the assay they used did not reveal such a function for HP6, suggesting that HP6 is not needed for heterochromatic transgene silencing (Greil et al. 2007).
Both HP1 and Umbrea contain a chromo shadow domain. This domain mediates homodimerization of HP1 (Brasher et al. 2000; Cowieson et al. 2000; Yamamoto and Sonoda 2003) and we have shown that this domain mediates heterodimerization between HP1 and Umbrea in vitro. This finding is supported by our immunoprecipitation assays. Hip and HP1 are co-precipitated with Umbrea, suggesting that all three proteins are associated in a protein complex in vivo. It should be noted that we have recently identified three HP1-binding interfaces in the Hip protein (Schwendemann et al. 2008). The presence of three binding interfaces in Hip implies a mode of cooperative binding suited to cross-linking of multiple chromo shadow domain-containing molecules like HP1 and Umbrea. It therefore cannot be ruled out that the in vivo interaction between Umbrea and HP1 is only indirect, mediated by the bridging protein Hip.
In agreement with this model, we find that Umbrea and HP1 use the same three binding modules within the Hip sequence. Both the chromo shadow domains of Umbrea and HP1 interact independently with the three binding interfaces of Hip. The interaction of the two different proteins with the same interaction modules in Hip supports our idea of a novel chromo shadow domain binding interface in Hip (Schwendemann et al. 2008). The Umbrea protein appears unique among other heterochromatin proteins since it is almost reduced to its chromo shadow domain. What might be the functional mechanism of a protein that consists of a single domain that is similar to the HP1 chromo shadow domain? In HP1 this domain provides the surface for the interaction with various other chromosomal proteins and displays the HP1 protein partner promiscuity (Eissenberg and Elgin 2000). In agreement with this, we have shown that Umbrea interacts in the same way with at least three proteins. Binding of Umbrea could block the binding surface of an interacting partner to prevent the interaction with other proteins.
It is known that HP1 is essential for heterochromatin localization of many proteins. We have recently shown that Hip binding to heterochromatin depends on HP1 (Schwendemann et al. 2008). In the study presented here we find that HP1 also serves as a binding platform for Umbrea. For this experiment we used HP1-deficient third-instar larvae and our result is not consistent with experiments of Greil et al. (2007). Greil et al. used RNAi to reduce HP1 levels. They found that chromosomal localization of HP6 (identical to Umbrea) was only subtly affected by HP1 depletion. We speculate that residual low amounts of HP1 after RNAi might be sufficient for Umbrea binding to chromatin.
Umbrea binding along the arms of polytene chromosomes seems to be unaffected by Hip depletion. Given the interaction of Umbrea with both Hip and HP1, it is likely that the Umbrea/HP1 interaction is sufficient to target Umbrea to chromatin in the absence of Hip. In turn, this seems to be the case for HP1 and Hip. Their binding appeared to be unaffected after RNAi-induced Umbrea depletion. In contrast, Umbrea association with chromocentre heterochromatin depends on Hip. Different requirements of Hip for Umbrea association with chromocentre and chromosomal arms suggest occurrence of heterochromatin protein complexes of different composition that differentially regulate the assembly of Umbrea-containing complexes.
Taking these findings together, we speculate that HP1 is a key player for heterochromatin targeting and serves as an essential binding platform for chromatin localization of Hip and Umbrea and many other proteins.
The Drosophila HOAP and HP1 proteins are stable components of telomeres and both proteins specifically interact with each other (Shareef et al. 2001; Badugu et al. 2003). Our cytogenetic studies revealed that Umbrea also localizes to telomeres. However, molecular and genetic analyses provide the evidence for existence of three distinct domains in distal regions of chromosomes: cap complex, which is assembled on the terminal DNA in a sequence-independent manner (Biessmann et al. 1990); the retrotransposon array of He-T-A/TAHRE/TART elements; and the subterminal TAS repeats (Biessmann and Mason 2003). Protein attachment to telomeric structures is not sufficient to establish that a protein is a component of the cap (Boivin et al. 2003; Andreyeva et al. 2005). Thus, from our cytogenetic analyses we cannot assign Umbrea localization to one of the three domains in telomere ends of polytene chromosomes. But given the association of HP1 and HOAP with the cap region (Fanti et al. 1998; Siriaco et al. 2002; Cenci et al. 2003) and the direct protein interaction of Umbrea with both HP1 and HOAP, we speculate that Umbrea also localizes to the cap region.
Mutations in Su(var)2-5 and cav cause extensive telomere–telomere fusions, indicating that the encoded proteins are essential for telomere stability and required for telomere capping and telomere fusion protection (Fanti et al. 1998; Shareef et al. 2001; Cenci et al. 2003; Perrini et al. 2004). We have shown that Umbrea physically interacts not only with HP1 but also with HOAP. Our cytogenetic studies revealed that Umbrea is a component of all telomeres. On the basis of these results, we expected a similar telomeric function for Umbrea. However, cytological analysis of larval brain cells displayed neither end-to-end attachments of metaphase chromosomes nor abnormal metaphase configurations. For analysis of mutant brain cells we used different approaches. The lethality of the umbrea P-element mutant line did not allow cytological analysis since homozygous animals die early during embryogenesis. In another approach we examined progeny of an umbrea specific RNAi line under the control of UAS in combination with different neuronal and ubiquitous Gal4 driver lines. Again, lethality precludes mutant characterization of metaphase chromosomes. Interestingly, the RNAi-induced depletion of Umbrea in salivary glands reveals a mutant phenotype. We found frequent telomere–telomere attachments in polytene nuclei. Given the localization of Umbrea at telomeres and the interaction of Umbrea with the telomere-associated proteins HP1 and HOAP, this result is not really surprising at first glance. However, the mechanisms by which telomeres attach to each other in polytene nuclei are not currently understood. It is speculated that mitotic and polytene chromosomes have different mechanisms of telomere protection (Cenci et al. 2005). In polytene chromosomes, telomere associations depend largely on the length of the retrotransposon arrays. On the other hand, in contrast to mitotic chromosomes, defects in the cap protein structure have not been shown to modify the frequencies of polytene telomere fusions (Fanti et al. 1998; Siriaco et al. 2002). Important differences observed between the polytene and mitotically dividing cells are speculated to be due to the fact that salivary gland differentiation and transition from mitotic divisions to endocycles takes place in early embryogenesis (Orr-Weaver 1994). In this respect, maternally contributed HP1 from heterozygous Su(var)2-5 mutants is still sufficient to suppress telomeric fusions (discussed in Andreyeva et al. 2005). However, we used a different RNAi-mediated approach to deplete Umbrea using the early embryonic driver line G61. Given the observed telomere–telomere fusion of polytene chromosomes, we speculate that the fusion potential depends critically on the onset of Umbrea protein reduction.
It is known that mutations in Su(var)2-5 cause both telomere fusion and telomere retrotransposon elongation (Savitsky et al. 2002). Ultimately, on the basis of our umbrea-specific RNAi analyses we cannot attribute the telomere fusion to defects in the protein cap structure or to the presence of excessive retrotransposon arrays. It might even be possible that Umbrea, like HP1, exhibits functions in both mechanisms. However, our results clearly indicate that umbrea elicits a phenotype similar to that observed in mutants in the HP1- and HOAP-encoding genes cav and Su(var)2-5 (Fanti et al. 1998; Cenci et al. 2003). We assume that Umbrea, together with HP1 and HOAP (and perhaps numerous additional proteins), forms a telomere-capping complex and is required for telomere function.
HP1 associates with heterochromatin, telomeres and multiple euchromatic sites. It is speculated that the different locations of HP1 are related to multiple different functions (reviewed in Fanti and Pimpinelli 2008). Umbrea is located not only at telomeres but also in the pericentric heterochromatin, at regions along the euchromatic arms and, interestingly, in the nucleolus. Given these different positions, we assume that the function of Umbrea is not limited to telomeres. The gene umbrea is essential for normal development since both the umbrea P-element mutant and RNAi depletion of Umbrea are lethal. Further studies are required for understanding the function of the chromo shadow domain protein Umbrea and its relationship with other heterochromatin binding proteins.
Abbreviations
- BDGR:
-
Berkeley Drosophila Genome Project
- cav :
-
caravaggio
- cd:
-
chromo domain
- cs:
-
chromo shadow domain
- dp :
-
dumpy
- GST:
-
glutathione-S-transferase
- Hip:
-
HP1-interacting protein
- HOAP:
-
HP1/ORC associated protein
- HP1:
-
heterochromatin protein 1
- ORC:
-
origin recognition complex
- PCR:
-
polymerase chain reaction
- PEV:
-
position effect variegation
- RNAi:
-
RNA interference
- TAS:
-
telomere-associated sequences
References
Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A (2004) TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol 21:1620–1624
Andreyeva EN, Belyaeva ES, Semeshin VF, Pokholkova GV, Zhimulev IF (2005) Three distinct chromatin domains in telomere ends of polytene chromosomes in rosophila melanogaster Tel mutants. J Cell Sci 118:5465–5477
Badugu R, Shareef MM, Kellum R (2003) Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J Biol Chem 278:34491–34498
Badugu R, Yoo Y, Singh PB, Kellum R (2005) Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma 113:370–384
Bannister AJ, Zegerman P, Partridge JF et al (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124
Biessmann H, Mason JM (2003) Telomerase-independent mechanisms of telomere elongation. Cell Mol Life Sci 60:2325–2333
Biessmann H, Carter SB, Mason JM (1990) Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA 87:1758–1761
Biessmann H, Champion LE, O’Hair M, Ikenaga K, Kasravi B, Mason JM (1992) Frequent transpositions of Drosophila melanogaster HeT-A transposable elements to receding chromosome ends. EMBO J 11:4459–4469
Boivin A, Gally C, Netter S, Anxolabehere D, Ronsseray S (2003) Telomeric associated sequences of Drosophila recruit polycomb-group proteins in vivo and can induce pairing-sensitive repression. Genetics 164:195–208
Brasher SV, Smith BO, Fogh RH et al (2000) The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J 19:1587–1597
Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M (2003) The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol 5:82–84
Cenci G, Ciapponi L, Gatti M (2005) The mechanism of telomere protection: a comparison between Drosophila and humans. Chromosoma 114:135–145
Cheah PY, Chia W, Yang X (2000) Jumeaux, a novel Drosophila winged-helix family protein, is required for generating asymmetric sibling neuronal cell fates. Development 127:3325–3335
Counter CM, Avilion AA, LeFeuvre CE et al (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. Embo J 11:1921–1929
Cowieson NP, Partridge JF, Allshire RC, McLaughlin PJ (2000) Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol 10:517–525
de Lange T (2002) Protection of mammalian telomeres. Oncogene 21:532–540
Dietzl G, Chen D, Schnorrer F et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156
Eissenberg JC, Elgin SC (2000) The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev 10:204–210
Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC (1990) Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci USA 87:9923–9927
Fanti L, Pimpinelli S (2008) HP1: a functionally multifaceted protein. Curr Opin Genet Dev 18(2):169–174, Epub 2008 Mar 10
Fanti L, Giovinazzo G, Berloco M, Pimpinelli S (1998) The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell 2:527–538
Fanti L, Berloco M, Piacentini L, Pimpinelli S (2003) Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: a cytological map of euchromatic HP1 binding sites. Genetica 117:135–147
Giot L, Bader JS, Brouwer C et al (2003) A protein interaction map of Drosophila melanogaster. Science 302:1727–1736
Greil F, de Wit E, Bussemaker HJ, van Steensel B (2007) HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. Embo J 26:741–751
Hiatt MA, Elde N, Vermaak D, Malik H (2007) Recycling the genome: Umbrea, a telomere-associated protein in Drosophila, arose from a duplicated HP1 protein. Dros Res Conf 48:325
James TC, Eissenberg JC, Craig C, Dietrich V, Hobson A, Elgin SC (1989) Distribution patterns of HP1: a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur J Cell Biol 50:170–180
Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120
Lehmann M, Korge G (1995) Ecdysone regulation of the Drosophila Sgs-4 gene is mediated by the synergistic action of ecdysone receptor and SEBP 3. EMBO J 14:716–726
Lehmann M, Korge G (1996) The fork head product directly specifies the tissue-specific hormone responsiveness of the Drosophila Sgs-4 gene. EMBO J 15:4825–4834
Li Y, Kirschmann DA, Wallrath LL (2002) Does heterochromatin protein 1 always follow code. Proc Natl Acad Sci USA 99(Suppl 4):16462–16469
Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC (2000) Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155:699–708
Maser RS, DePinho RA (2002) Connecting chromosomes, crisis, and cancer. Science 297:565569
Orr-Weaver TL (1994) Developmental modification of the Drosophila cell cycle. Trends Genet 10:321–327
Pak DT, Pflumm M, Chesnokov I et al (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311–323
Perrini B, Piacentini L, Fanti L et al (2004) HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell 15:467–476
Savitsky M, Kravchuk O, Melnikova L, Georgiev P (2002) Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol 22:3204–3218
Schwendemann A, Matkovic T, Linke C, Klebes A, Hofmann A, Korge G (2008) Hip, an HP1-interacting protein, is a haplo- and triplo-suppressor of position effect variegation. Proc Natl Acad Sci USA 105:204–209
Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R (2001) Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell 12:1671–1685
Sheen FM, Levis RW (1994) Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA 91:12510–12514
Siegmund T, Korge G (2001) Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol 431:481–491
Singh PB, Georgatos SD (2002) HP1: facts, open questions, and speculation. J Struct Biol 140:10–16
Siriaco GM, Cenci G, Haoudi A et al (2002) Telomere elongation (Tel), a new mutation in Drosophila melanogaster that produces long telomeres. Genetics 160:235–445
Smothers JF, Henikoff S (2001) The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol 21:2555–2569
Strodicke M, Karberg S, Korge G (2000) Domina (Dom), a new Drosophila member of the FKH/WH gene family, affects morphogenesis and is a suppressor of position-effect variegation. Mech Dev 96:67–78
Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC (2000) The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc Natl Acad Sci USA 97:5340–5345
van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92:401–413
Yamamoto K, Sonoda M (2003) Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase SUV39H1. Biochem Biophys Res Commun 301:287–292
Acknowledgements
We thank S. C. R. Elgin for providing anti-HP1 antibody, G. Reuter for fly stocks, and M. Brünner for great technical assistance. This work was financially supported by a pilot project grant of the Freie Universität Berlin to A. S.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Walther Traut.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Joppich, C., Scholz, S., Korge, G. et al. Umbrea, a chromo shadow domain protein in Drosophila melanogaster heterochromatin, interacts with Hip, HP1 and HOAP. Chromosome Res 17, 19–36 (2009). https://doi.org/10.1007/s10577-008-9002-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-008-9002-1