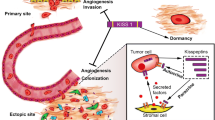
Abstract
Present therapeutic approaches do not effectively target metastatic cancers, often limited by their inability to eliminate already-seeded non-proliferative, growth-arrested, or therapy-resistant tumor cells. Devising effective approaches targeting dormant tumor cells has been a focus of cancer clinicians for decades. However, progress has been limited due to limited understanding of the tumor dormancy process. Studies on tumor dormancy have picked up pace and have resulted in the identification of several regulators. This review focuses on KISS1, a metastasis suppressor gene that suppresses metastasis by keeping tumor cells in a state of dormancy at ectopic sites. The review explores mechanistic insights of KISS1 and discusses its potential application as a therapeutic against metastatic cancers by eliminating quiescent cells or inducing long-term dormancy in tumor cells.

Similar content being viewed by others
References
Welch, D. R., & Hurst, D. R. (2019). Defining the hallmarks of metastasis. Cancer Research, 79(12), 3011–3027. https://doi.org/10.1158/0008-5472.CAN-19-0458
Fidler, I. J. (1970). Metastasis: Quantitative analysis of distribution and fate of tumor emboli labeled with 125l–5-lodo-2’-deoxyuridine. Journal of the National Cancer Institute, 45(4), 773–782. https://doi.org/10.1093/jnci/45.4.773
Moore, N., & Lyle, S. (2011). Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. Journal of Oncology, 2011, 1–11. https://doi.org/10.1155/2011/396076
Recasens, A., & Munoz, L. (2019). Targeting cancer cell dormancy. Trends in Pharmacological Sciences, 40(2), 128–141. https://doi.org/10.1016/j.tips.2018.12.004
Tang, D. G. (2012). Understanding cancer stem cell heterogeneity and plasticity. Cell Research, 22(3), 457–472. https://doi.org/10.1038/cr.2012.13
Phan, T. G., & Croucher, P. I. (2020). The dormant cancer cell life cycle. Nature Reviews Cancer, 20(7), 398–411. https://doi.org/10.1038/s41568-020-0263-0
Davis, J. E. J., Kirk, J., Ji, Y., & Tang, D. G. (2019). Tumor dormancy and slow-cycling cancer cells. Advances in Experimental Medicine and Biology, 1164, 199–206. https://doi.org/10.1007/978-3-030-22254-3_15
Bragado, P., Sosa, M. S., Keely, P., Condeelis, J., & Aguirre-Ghiso, J. A. (2012). Microenvironments dictating tumor cell dormancy. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer, 195, 25–39. https://doi.org/10.1007/978-3-642-28160-0_3
Gomis, R. R., & Gawrzak, S. (2017). Tumor cell dormancy. Molecular oncology, 11(1), 62–78. https://doi.org/10.1016/j.molonc.2016.09.009
Hedley, B. D., Winquist, E., & Chambers, A. F. (2004). Therapeutic targets for antimetastatic therapy. Expert opinion on therapeutic targets, 8(6), 527–536. https://doi.org/10.1517/14728222.8.6.527
Goss, P. E., & Chambers, A. F. (2010). Does tumour dormancy offer a therapeutic target? Nature reviews. Cancer, 10(12), 871–877. https://doi.org/10.1038/nrc2933
Neophytou, C. M., Kyriakou, T.-C., & Papageorgis, P. (2019). Mechanisms of metastatic tumor dormancy and implications for cancer therapy. International journal of molecular sciences, 20(24). https://doi.org/10.3390/ijms20246158
Saleh, T., Bloukh, S., Carpenter, V. J., Alwohoush, E., Bakeer, J., Darwish, S., … Gewirtz, D. A. (2020). Therapy-induced senescence: An “old” friend becomes the enemy. Cancers, 12(4). https://doi.org/10.3390/cancers12040822
Neophytou, C., Boutsikos, P., & Papageorgis, P. (2018). Molecular mechanisms and emerging therapeutic targets of triple-negative breast cancer metastasis. Frontiers in oncology, 8, 31. https://doi.org/10.3389/fonc.2018.00031
Iiizumi, M., Liu, W., Pai, S. K., Furuta, E., & Watabe, K. (2008). Drug development against metastasis-related genes and their pathways: A rationale for cancer therapy. Biochimica et biophysica acta, 1786(2), 87–104. https://doi.org/10.1016/j.bbcan.2008.07.002
Hurst, D. R., & Welch, D. R. (2011). Metastasis suppressor genes. At the interface between the environment and tumor cell growth. International Review of Cell and Molecular Biology, 286, 107–180. https://doi.org/10.1016/B978-0-12-385859-7.00003-3
Steeg, P. S., Ouatas, T., Halverson, D., Palmieri, D., & Salerno, M. (2003). Metastasis suppressor genes: Basic biology and potential clinical use. Clinical breast cancer, 4(1), 51–62. https://doi.org/10.3816/cbc.2003.n.012
Smith, S. C., & Theodorescu, D. (2009). Learning therapeutic lessons from metastasis suppressor proteins. Nature Reviews Cancer, 9, 253–264. https://doi.org/10.1038/nrc2594
Park, K. C., Paluncic, J., Kovacevic, Z., & Richardson, D. R. (2020). Pharmacological targeting and the diverse functions of the metastasis suppressor, NDRG1, in cancer. Free Radical Biology and Medicine, 157, 154–175. https://doi.org/10.1016/j.freeradbiomed.2019.05.020
Kauffman, E. C., Robinson, V. L., Stadler, W. M., Sokoloff, M. H., & Rinker-Schaeffer, C. W. (2003). Metastasis suppression: The evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. Journal of Urology, 169(3), 1122–1133. https://doi.org/10.1097/01.ju.0000051580.89109.4b
Bohl, C. R., Harihar, S., Denning, W. L., Sharma, R., & Welch, D. R. (2014). Metastasis suppressors in breast cancers: Mechanistic insights and clinical potential. Journal of Molecular Medicine, 92(1), 13–30. https://doi.org/10.1007/s00109-013-1109-y
Beck, B. H., & Welch, D. R. (2010). The KISS1 metastasis suppressor: A good night kiss for disseminated cancer cells. European Journal of Cancer, 46(7), 1283–1289. https://doi.org/10.1016/j.ejca.2010.02.023
Chen, X., Xu, Z., & Wang, Y. (2011). Recent advances in breast cancer metastasis suppressor 1. The International journal of biological markers, 26(1), 1–8. https://doi.org/10.5301/jbm.2011.6267
Hedley, B. D., & Chambers, A. F. (2009). Tumor dormancy and metastasis. Advances in cancer research, 102, 67–101. https://doi.org/10.1016/S0065-230X(09)02003-X
Osisami, M., & Keller, E. T. (2013). Mechanisms of metastatic tumor dormancy. Journal of clinical medicine, 2(3), 136–150. https://doi.org/10.3390/jcm2030136
Horak, C. E., Lee, J. H., Marshall, J. C., Shreeve, S. M., & Steeg, P. S. (2008). The role of metastasis suppressor genes in metastatic dormancy. APMIS, 116(7–8), 586–601. https://doi.org/10.1111/j.1600-0463.2008.01027.x
Harihar, S., Ray, S., Narayanan, S., Santhoshkumar, A., Ly, T., & Welch, D. R. (2020). Role of the tumor microenvironment in regulating the anti-metastatic effect of KISS1. Clinical and Experimental Metastasis, 37(2). https://doi.org/10.1007/s10585-020-10030-6
Ly, T., Harihar, S., & Welch, D. R. (2020). KISS1 in metastatic cancer research and treatment: Potential and paradoxes. Cancer and Metastasis Reviews, 39, 739–754. https://doi.org/10.1007/s10555-020-09868-9
Crist, S. B., & Ghajar, C. M. (2021). When a house is not a home: A survey of antimetastatic niches and potential mechanisms of disseminated tumor cell suppression. Annual Review of Pathology: Mechanisms of Disease, 16, 409–432. https://doi.org/10.1146/annurev-pathmechdis-012419-032647
Kang, Y., & Pantel, K. (2013). Tumor cell dissemination: Emerging biological insights from animal models and cancer patients. Cancer Cell, 23(5), 573–581. https://doi.org/10.1016/j.ccr.2013.04.017
Wikman, H., Vessella, R., & Pantel, K. (2008). Cancer micrometastasis and tumour dormancy. APMIS : Acta pathologica, microbiologica, et immunologica Scandinavica, 116(7–8), 754–770. https://doi.org/10.1111/j.1600-0463.2008.01033.x
Izraely, S., & Witz, I. P. (2021). Site-specific metastasis: A cooperation between cancer cells and the metastatic microenvironment. International journal of cancer, 148(6), 1308–1322. https://doi.org/10.1002/ijc.33247
Grzelak, C. A., & Ghajar, C. M. (2017). Metastasis “systems” biology: How are macro-environmental signals transmitted into microenvironmental cues for disseminated tumor cells? Current opinion in cell biology, 48, 79–86. https://doi.org/10.1016/j.ceb.2017.06.002
Mirzayans, R., Andrais, B., & Murray, D. (2018). Roles of polyploid/multinucleated giant cancer cells in metastasis and disease relapse following anticancer treatment. Cancers, 10(4). https://doi.org/10.3390/cancers10040118
Sceneay, J., Smyth, M. J., & Möller, A. (2013). The pre-metastatic niche: Finding common ground. Cancer metastasis reviews, 32(3–4), 449–464. https://doi.org/10.1007/s10555-013-9420-1
Liu, Y., & Cao, X. (2016). Characteristics and significance of the pre-metastatic niche. Cancer Cell, 30(5), 668–681. https://doi.org/10.1016/j.ccell.2016.09.011
Aguirre-Ghiso, J. A., & Sosa, M. S. (2018). Emerging topics on disseminated cancer cell dormancy and the paradigm of metastasis. Annual Review of Cancer Biology, 2, 377–393. https://doi.org/10.1146/annurev-cancerbio-030617-050446
Araos, J., Sleeman, J. P., & Garvalov, B. K. (2018). The role of hypoxic signalling in metastasis: Towards translating knowledge of basic biology into novel anti-tumour strategies. Clinical & experimental metastasis, 35(7), 563–599. https://doi.org/10.1007/s10585-018-9930-x
Butturini, E., Carcereri de Prati, A., Boriero, D., & Mariotto, S. (2019). Tumor dormancy and interplay with hypoxic tumor microenvironment. International journal of molecular sciences, 20(17). https://doi.org/10.3390/ijms20174305
Walker, N. D., Patel, J., Munoz, J. L., Hu, M., Guiro, K., Sinha, G., & Rameshwar, P. (2016). The bone marrow niche in support of breast cancer dormancy. Cancer letters, 380(1), 263–271. https://doi.org/10.1016/j.canlet.2015.10.033
Attaran, S., & Bissell, M. J. (2022). The role of tumor microenvironment and exosomes in dormancy and relapse. Seminars in cancer biology, 78, 35–44. https://doi.org/10.1016/j.semcancer.2021.09.008
Hernández-Barranco, A., Nogués, L., & Peinado, H. (2021). Could extracellular vesicles contribute to generation or awakening of “sleepy” metastatic niches? Frontiers in cell and developmental biology, 9, 625221. https://doi.org/10.3389/fcell.2021.625221
Patel, S. A., Meyer, J. R., Greco, S. J., Corcoran, K. E., Bryan, M., & Rameshwar, P. (2010). Mesenchymal stem cells protect breast cancer cells through regulatory T cells: Role of mesenchymal stem cell-derived TGF-β. The Journal of Immunology, 184(10), 5885–5894. https://doi.org/10.4049/jimmunol.0903143
Roodman, G. D. (2004). Mechanisms of bone metastasis. New England Journal of Medicine, 350(16), 1655–1664. https://doi.org/10.1056/nejmra030831
Psaila, B., & Lyden, D. (2009). The metastatic niche: Adapting the foreign soil. Nature Reviews Cancer, 9, 285–293. https://doi.org/10.1038/nrc2621
Dianat-Moghadam, H., Azizi, M., Eslami-S, Z., Cortés-Hernández, L. E., Heidarifard, M., Nouri, M., & Alix-Panabières, C. (2020). The role of circulating tumor cells in the metastatic cascade: Biology, technical challenges, and clinical relevance. Cancers, 12(4). https://doi.org/10.3390/cancers12040867
Joosse, S. A., Gorges, T. M., & Pantel, K. (2015). Biology, detection, and clinical implications of circulating tumor cells. EMBO molecular medicine, 7(1), 1–11. https://doi.org/10.15252/emmm.201303698
Ramamoorthi, G., Kodumudi, K., Gallen, C., Zachariah, N. N., Basu, A., Albert, G., … Czerniecki, B. J. (2022). Disseminated cancer cells in breast cancer: Mechanism of dissemination and dormancy and emerging insights on therapeutic opportunities. Seminars in cancer biology, 78, 78–89. https://doi.org/10.1016/j.semcancer.2021.02.004
Esposito, M., & Kang, Y. (2014). Targeting tumor-stromal interactions in bone metastasis. Pharmacology & therapeutics, 141(2), 222–233. https://doi.org/10.1016/j.pharmthera.2013.10.006
Aguado, B. A., Bushnell, G. G., Rao, S. S., Jeruss, J. S., & Shea, L. D. (2017). Engineering the pre-metastatic niche. Nature Biomedical Engineering, 1(0077). https://doi.org/10.1038/s41551-017-0077
Lim, A. R., & Ghajar, C. M. (2022). Thorny ground, rocky soil: Tissue-specific mechanisms of tumor dormancy and relapse. Seminars in cancer biology, 78, 104–123. https://doi.org/10.1016/j.semcancer.2021.05.007
Kamińska, K., Szczylik, C., & Bielecka, Z. F. (2015). The role of the cell-cell interactions in cancer progression. Journal of Cellular and Molecular Medicine, 19(2), 283–296. https://doi.org/10.1111/jcmm.12408
De Wever, O., Van Bockstal, M., Mareel, M., Hendrix, A., & Bracke, M. (2014). Carcinoma-associated fibroblasts provide operational flexibility in metastasis. Seminars in cancer biology, 25, 33–46. https://doi.org/10.1016/j.semcancer.2013.12.009
Kwa, M. Q., Herum, K. M., & Brakebusch, C. (2019). Cancer-associated fibroblasts: How do they contribute to metastasis? Clinical and Experimental Metastasis, 36, 71–86. https://doi.org/10.1007/s10585-019-09959-0
Raz, Y., Cohen, N., Shani, O., Bell, R. E., Novitskiy, S. V., Abramovitz, L., … Erez, N. (2018). Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. Journal of Experimental Medicine, 215(12), 3075–3093. https://doi.org/10.1084/jem.20180818
Yeo, S. Y., Lee, K. W., Shin, D., An, S., Cho, K. H., & Kim, S. H. (2018). A positive feedback loop bi-stably activates fibroblasts. Nature Communications, 9(1). https://doi.org/10.1038/s41467-018-05274-6
Psaila, B., Kaplan, R. N., Port, E. R., & Lyden, D. (2006). Priming the “soil” for breast cancer metastasis: The pre-metastatic niche. Breast disease, 26, 65–74. https://doi.org/10.3233/bd-2007-26106
Ping, Y. F., Yao, X. H., Jiang, J. Y., Zhao, L. T., Yu, S. C., Jiang, T., … Bian, X. W. (n.d.). The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling. The Journal of pathology, 224(3), 344–354. https://doi.org/10.1002/path.2908
Potente, M., & Carmeliet, P. (2017). The link between angiogenesis and endothelial metabolism. Annual Review of Physiology. Annual Reviews Inc. https://doi.org/10.1146/annurev-physiol-021115-105134
Wilde, L., Roche, M., Domingo-Vidal, M., Tanson, K., Philp, N., Curry, J., & Martinez-Outschoorn, U. (2017). Metabolic coupling and the Reverse Warburg Effect in cancer: Implications for novel biomarker and anticancer agent development. Seminars in Oncology, 44(3), 198–203. https://doi.org/10.1053/j.seminoncol.2017.10.004
Bott, A. J., Maimouni, S., & Zong, W. X. (2019). The pleiotropic effects of glutamine metabolism in cancer. Cancers, 11(6). https://doi.org/10.3390/cancers11060770
Yan, S., Vandewalle, N., De Beule, N., Faict, S., Maes, K., De Bruyne, E., … De Veirman, K. (2019). AXL receptor tyrosine kinase as a therapeutic target in hematological malignancies: Focus on multiple myeloma. Cancers, 11(11). https://doi.org/10.3390/cancers11111727
Yumoto, K., Eber, M. R., & Wang, J. (2016). Axl is required for TGF-β2-induced dormancy of prostate cancer cells in the bone marrow. Sci Rep, 6(36520). https://doi.org/10.1038/srep36520
Prunier, C., Baker, D., ten Dijke, P., & Ritsma, L. (2019). TGF-β family signaling pathways in cellular dormancy. Trends in cancer, 5(1), 66–78. https://doi.org/10.1016/J.TRECAN.2018.10.010
Buijs, J. T., Henriquez, N. V., van Overveld, P. G. M., van der Horst, G., ten Dijke, P., & van der Pluijm, G. (2007). TGF-beta and BMP7 interactions in tumour progression and bone metastasis. Clinical & experimental metastasis, 24(8), 609–617. https://doi.org/10.1007/s10585-007-9118-2
Bragado, P., Estrada, Y., & Parikh, F. (2013). TGF-β2 dictates disseminated tumour cell fate in target organs through TGF-β-RIII and p38α/β signalling. Nature Cell Biology, 15(11), 1351–1361. https://doi.org/10.1038/ncb2861
Kovacevic, Z., Chikhani, S., Lui, G. Y. L., Sivagurunathan, S., & Richardson, D. R. (2013). The iron-regulated metastasis suppressor NDRG1 targets NEDD4L, PTEN, and SMAD4 and inhibits the PI3K and ras signaling pathways. Antioxidants and Redox Signaling, 18(8), 874–887. https://doi.org/10.1089/ars.2011.4273
Sun, J., Zhang, D., Bae, D. H., Sahni, S., Jansson, P., Zheng, Y., … Richardson, D. R. (2013). Metastasis suppressor, NDRG1, mediates its activity through signaling pathways and molecular motors. Carcinogenesis, 34, 1943–1954. https://doi.org/10.1093/carcin/bgt163
Patel, S., Alam, A., Pant, R., & Chattopadhyay, S. (2019). Wnt signaling and its significance within the tumor microenvironment: Novel therapeutic insights. Front Immunol, 10(2872). https://doi.org/10.3389/fimmu.2019.02872
Carreira-Barbosa, F., & Nunes, S. C. (2020). Wnt signaling: Paths for cancer progression. Advances in experimental medicine and biology, 1219, 189–202. https://doi.org/10.1007/978-3-030-34025-4_10
Ranganathan, A. C., Adam, A. P., Zhang, L., & Aguirre-Ghiso, J. A. (2006). Tumor cell dormancy induced by p38SAPK and ER-stress signaling: An adaptive advantage for metastatic cells? Cancer biology & therapy, 5(7), 729–735. https://doi.org/10.4161/cbt.5.7.2968
Roux, P. P., & Blenis, J. (2004). ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiology and Molecular Biology Reviews, 68(2), 320–344. https://doi.org/10.1128/MMBR.68.2.320-344.2004
Jahanban-Esfahlan, R., Seidi, K., Manjili, M. H., Jahanban-Esfahlan, A., Javaheri, T., & Zare, P. (2019). Tumor cell dormancy: Threat or opportunity in the fight against cancer. Cancers, 11(8), 1207. https://doi.org/10.3390/cancers11081207
Kudaravalli, S., den Hollander, P., & Mani, S. A. (2022). Role of p38 MAP kinase in cancer stem cells and metastasis. Oncogene, 41(23), 3177–3185. https://doi.org/10.1038/s41388-022-02329-3
Sosa, M. S., Parikh, F., & Maia, A. G. (2015). NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes. Nature Communications, 6(1). https://doi.org/10.1038/ncomms7170
Goddard, E. T., Bozic, I., Riddell, S. R., & Ghajar, C. M. (2018). Dormant tumour cells, their niches and the influence of immunity. Nature cell biology, 20(11), 1240–1249. https://doi.org/10.1038/s41556-018-0214-0
Massagué, J., & Ganesh, K. (2021). Metastasis-initiating cells and ecosystems. Cancer discovery, 11(4), 971–994. https://doi.org/10.1158/2159-8290.CD-21-0010
Wang, H.-F., Wang, S.-S., Huang, M.-C., Liang, X.-H., Tang, Y.-J., & Tang, Y.-L. (2019). Targeting immune-mediated dormancy: A promising treatment of cancer. Frontiers in oncology, 9, 498. https://doi.org/10.3389/fonc.2019.00498
Lopes, N., & Vivier, E. (2021). Natural killer cells lull tumors into dormancy. Nature, 594(7864), 501–502. https://doi.org/10.1038/d41586-021-01381-5
Steel, J. C., Waldmann, T. A., & Morris, J. C. (2012). Interleukin-15 biology and its therapeutic implications in cancer. Trends in Pharmacological Sciences, 33(1), 35–41. https://doi.org/10.1016/j.tips.2011.09.004
Barra, N. G., Chew, M. v, Reid, S., & Ashkar, A. A. (2012). Interleukin-15 treatment induces weight loss independent of lymphocytes. PLoS One, 7(6). https://doi.org/10.1371/journal.pone.0039553
Shi Y Riese DJ 2nd, S. J. (2020). The role of the CXCL12/CXCR4/CXCR7 chemokine axis in cancer. Frontiers Pharmacology, 11(574667). https://doi.org/10.3389/fphar.2020.574667
Farrar, J. D., Katz, K. H., Windsor, J., Thrush, G., Scheuermann, R. H., Uhr, J. W., & Street, N. E. (1999). Cancer dormancy. VII. A regulatory role for CD8+ T cells and IFN-gamma in establishing and maintaining the tumor-dormant state. Journal of immunology, 162(5), 2842–2849.
Mayhew, V., Omokehinde, T., & Johnson, R. W. (2020). Tumor dormancy in bone. Cancer reports (Hoboken, N.J.), 3(1), e1156. https://doi.org/10.1002/cnr2.1156
Lan, Q., Peyvandi, S., & Duffey, N. (2019). Type I interferon/IRF7 axis instigates chemotherapy-induced immunological dormancy in breast cancer. Oncogene, 38(15), 2814–2829. https://doi.org/10.1038/s41388-018-0624-2
Bidwell, B. N., Slaney, C. Y., & Withana, N. P. (2012). Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nature Medicine, 18(8), 1224–1231. https://doi.org/10.1038/nm.2830
Ostroumov, D., Fekete-Drimusz, N., Saborowski, M., Kühnel, F., & Woller, N. (2018). CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cellular and Molecular Life Sciences, 75(4), 689–713. https://doi.org/10.1007/s00018-017-2686-7
Flies, D. B., Sandler, B. J., Sznol, M., & Chen, L. (2011). Blockade of the B7–H1/PD-1 pathway for cancer immunotherapy. The Yale Journal of Biology and Medicine, 84(4), 409–421.
Topalian, S. L., Drake, C. G., & Pardoll, D. M. (2012). Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Current Opinion in Immunology, 24(2), 207–212. https://doi.org/10.1016/j.coi.2011.12.009
Jorgovanovic, D., Song, M., Wang, L., & Zhang, Y. (2020). Roles of IFN-γ in tumor progression and regression: A review. Biomark Research, 8(1). https://doi.org/10.1186/s40364-020-00228-x
Burke, J. D., & Young, H. A. (2019). IFN-γ: A cytokine at the right time, is in the right place. Seminars Immunology, 43(101280). https://doi.org/10.1016/j.smim.2019.05.002
Albrengues, J., Shields, M. A., Ng, D., Park, C. G., Ambrico, A., Poindexter, M. E., … Egeblad, M. (2018). Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science, 361(6409). https://doi.org/10.1126/science.aao4227
Tohme, S., Yazdani, H. O., Al-Khafaji, A. B., Chidi, A. P., Loughran, P., Mowen, K., … Tsung, A. (2016). Neutrophil extracellular traps promote the development and progression of liver metastases after surgical stress. Cancer Research, 76(6), 1367–1380. https://doi.org/10.1158/0008-5472.CAN-15-1591
Khan, I., & Steeg, P. S. (2018). Metastasis suppressors: Functional pathways. Laboratory Investigation, 98, 198–210. https://doi.org/10.1038/labinvest.2017.104
Stafford, L. J., Vaidya, K. S., & Welch, D. R. (2008). Metastasis suppressors genes in cancer. International Journal of Biochemistry and Cell Biology, 40(5), 874–891. https://doi.org/10.1016/j.biocel.2007.12.016
Nash, K. T., Phadke, P. A., Navenot, J. M., Hurst, D. R., Accavitti-Loper, M. A., Sztul, E., … Welch, D. R. (2007). Requirement of KISS1 secretion for multiple organ metastasis suppression and maintenance of tumor dormancy [published correction appears in J Natl Cancer Inst. Journal of National Cancer Institute, 99(4), 309–321. https://doi.org/10.1093/jnci/djk053
Steeg, P. S., Bevilacqua, G., Pozzatti, R., Liotta, L. A., & Sobel, M. E. (1988). Altered expression of NM23, a gene associated with low tumor metastatic potential, during adenovirus 2 Ela inhibition of experimental metastasis. Cancer Research, 48(22), 6550–6554.
Welch, D. R., Chen, P., Miele, M. E., McGary, C. T., Bower, J. M., Stanbridge, E. J., & Weissman, B. E. (1994). Microcell-mediated transfer of chromosome 6 into metastatic human C8161 melanoma cells suppresses metastasis but does not inhibit tumorigenicity. Oncogene, 9(1), 255–262.
Goldberg, S. F., Miele, M. E., Hatta, N., Takata, M., Paquette-Straub, C., Freedman, L. P., & Welch, D. R. (2003). Melanoma metastasis suppression by chromosome 6: Evidence for a pathway regulated by CRSP3 and TXNIP. Cancer Research, 63(2), 432–440.
Cebrian, V., Fierro, M., Orenes-Piero, E., Grau, L., Moya, P., Ecke, T., … Sánchez-Carbayo, M. (2011). KISS1 methylation and expression as tumor stratification biomarkers and clinical outcome prognosticators for bladder cancer patients. American Journal of Pathology, 179(2), 540–546. https://doi.org/10.1016/j.ajpath.2011.05.009
Lee, J. H., & Welch, D. R. (1997). Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Research, 57(12), 2384–2387.
Cvetković, D., Babwah, A. V., & Bhattacharya, M. (2013). Kisspeptin/KISSIR system in breast cancer. Journal of Cancer, 4(8), 653–661. https://doi.org/10.7150/jca.7626
McNally, L. R., Welch, D. R., Beck, B. H., Stafford, L. J., Long, J. W., Sellers, J. C., … Buchsbaum, D. J. (2010). KISS1 over-expression suppresses metastasis of pancreatic adenocarcinoma in a xenograft mouse model. Clinical and Experimental Metastasis, 27(8), 591–600. https://doi.org/10.1007/s10585-010-9349-5
Jiang, Y., Berk, M., Singh, L. S., Tan, H., Yin, L., Powell, C. T., & Xu, Y. (2005). KiSS1 suppresses metastasis in human ovarian cancer via inhibition of protein kinase C alpha. Clinical and Experimental Metastasis, 22(5), 369–376. https://doi.org/10.1007/s10585-005-8186-4
Okugawa, Y., Inoue, Y., Tanaka, K., Toiyama, Y., Shimura, T., Okigami, M., … M., K. (2013). Loss of the metastasis suppressor gene KiSS1 is associated with lymph node metastasis and poor prognosis in human colorectal cancer. Oncology Reports, 30(3), 1449–1454. https://doi.org/10.3892/or.2013.2558
Kostakis, I. D., Agrogiannis, G., Vaiopoulos, A. G., Mylona, E., Patsouris, E., Kouraklis, G., & Koutsilieris, M. (2015). A clinicopathological analysis of KISS1 and KISS1R expression in colorectal cancer. APMIS, 123(7), 629–637. https://doi.org/10.1111/apm.12397
Kostakis, I. D., Agrogiannis, G., Vaiopoulos, A. G., Mylona, E., Patsouris, E., Kouraklis, G., & Koutsilieris, M. (2018). KISS1 and KISS1R expression in gastric cancer. Journal of B.U.O.N, 23(3), 598–603.
Lee, J. H., Miele, M. E., Hicks, D. J., Phillips, K. K., Trent, J. M., Weissman, B. E., & Welch, D. R. (1996). KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. Journal of the National Cancer Institute, 88(23), 1731–1737. https://doi.org/10.1093/jnci/88.23.1731
Zhang, Y., Tang, Y. J., Li, Z. H., Pan, F., Huang, K., & Xu, G. H. (2013). KiSS1 inhibits growth and invasion of osteosarcoma cells through inhibition of the MAPK pathway. European Journal of Histochemistry, 57(4), 200–205. https://doi.org/10.4081/ejh.2013.e30
Zheng, S., Chang, Y., Hodges, K. B., Sun, Y., Ma, X., Xueyi, … Cheng, L. (2010). Expression of KISS1 and MMP-9 in non-small cell lung cancer and their relations to metastasis and survival.Anticancer Research, 30(3), 713–718.
Sun, Y. B., & Xu, S. (2013). Expression of KISS1 and KISS1R (GPR54) may be used as favorable prognostic markers for patients with non-small cell lung cancer. International Journal of Oncology, 43(2), 521–530. https://doi.org/10.3892/ijo.2013.1967
Schmidt, E., Haase, M., Ziegler, E., Emons, G., & Gründker, C. (2014). Kisspeptin-10 inhibits stromal-derived factor 1-induced invasion of human endometrial cancer cells. International Journal of Gynecological Cancer, 24(2), 210–217. https://doi.org/10.1097/IGC.0000000000000050
Martins, C. M. O., Fernandes, B. F., Antecka, E., Di Cesare, S., Mansure, J. J. C., Marshall, J. C., & Burnier, M. N. (2008). Expression of the metastasis suppressor gene KISS1 in uveal melanoma. Eye, 22(5), 707–711. https://doi.org/10.1038/sj.eye.6703090
Jiffar, T., Yilmaz, T., Lee, J., Hanna, E., El-Naggar, A., Yu, D., … Kupferman, M. E. (2011). KiSS1 mediates platinum sensitivity and metastasis suppression in head and neck squamous cell carcinoma. Oncogene, 30(28), 3163–3173. https://doi.org/10.1038/onc.2011.39
Chen, Y., Zhang, C., Chen, J., Zhang, B., Zhang, H., Yang, X., … Wu, Q. (2018). Expression of transcription factor 21 (TCF21) and upregulation its level inhibits invasion and metastasis in esophageal squamous cell carcinoma. Medical Science Monitor, 24, 4128–4136. https://doi.org/10.12659/msm.909138
Ikeguchi, M., Hirooka, Y., & Kaibara, N. (2003). Quantitative reverse transcriptase polymerase chain reaction analysis for KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in hepatocellular carcinoma. Journal of Cancer Research and Clinical Oncology, 129, 531–535. https://doi.org/10.1007/s00432-003-0469-z
Zang, S., Liu, J. F., Wang, B., Gao, L., & Huang, A. (2009). Expression of KiSS-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anatomical Record, 292(8), 1128–1134. https://doi.org/10.1002/ar.20950
Goertzen, C. G., Dragan, M., Turley, E., Babwah, A. V., & Bhattacharya, M. (2016). KISS1R signaling promotes invadopodia formation in human breast cancer cell via β-arrestin2/ERK. Cellular Signalling, 28(3), 165–176. https://doi.org/10.1016/j.cellsig.2015.12.010
Tian, J., Al-Odaini, A. A., Wang, Y., Korah, J., Dai, M., Xiao, L., … Lebrun, J. J. (2018). KiSS1 gene as a novel mediator of TGFβ-mediated cell invasion in triple negative breast cancer. Cellular Signalling, 42, 1–10. https://doi.org/10.1016/j.cellsig.2017.10.002
Kim, C.-W., Lee, H. K., Nam, M.-W., Lee, G., & Choi, K.-C. (2022). The role of KiSS1 gene on the growth and migration of prostate cancer and the underlying molecular mechanisms. Life sciences, 310, 121009. https://doi.org/10.1016/j.lfs.2022.121009
Marot, D., Bieche, I., Aumas, C., Esselin, S., Bouquet, C., Vacher, S., … De Roux, N. (2007). High tumoral levels of Kiss1 and G-protein-coupled receptor 54 expression are correlated with poor prognosis of estrogen receptor-positive breast tumors. Endocrine-Related Cancer, 14(3), 691–702. https://doi.org/10.1677/ERC-07-0012
Lee, D. K., Nguyen, T., O’Neill, G. P., Cheng, R., Liu, Y., Howard, A. D., … O’Dowd, B. F. (1999). Discovery of a receptor related to the galanin receptors. FEBS Letters, 446(1), 103–107. https://doi.org/10.1016/S0014-5793(99)00009-5
Ohtaki, T., Shintani, Y., Honda, S., Matsumoto, H., Hori, A., Kanehashi, K., … Fujino, M. (2001). Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature, 411(6837), 613–617. https://doi.org/10.1038/35079135
Muir, A. I., Chamberlain, L., Elshourbagy, N. A., Michalovich, D., Moore, D. J., Calamari, A., … Harrison, D. C. (2001). AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. Journal of Biological Chemistry, 276, 28969–28975. https://doi.org/10.1074/jbc.M102743200
Kotani, M., Detheux, M., Vandenbogaerde, A., Communi, D., Vanderwinden, J. M., le Poul, E., … Parmentier, M. (2001). The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. Journal of Biological Chemistry, 276(37), 34631–34636. https://doi.org/10.1074/jbc.M104847200
Seminara, S. B. (2005). Metastin and its G protein-coupled receptor, GPR54: Critical pathway modulating GnRH secretion. Frontiers in neuroendocrinology, 26(3–4), 131–138. https://doi.org/10.1016/j.yfrne.2005.10.001
Nimri, R., Lebenthal, Y., Lazar, L., Chevrier, L., Phillip, M., Bar, M., … Gat-Yablonski, G. (2011). A novel loss-of-function mutation in GPR54/KISS1R leads to hypogonadotropic hypogonadism in a highly consanguineous family. Journal of Clinical Endocrinology Metabolism, 96(3), E536–45. https://doi.org/10.1210/jc.2010-1676
Carel, J.-C., Chaussain, J.-L., Milgrom, E., de Roux, N., Genin, E., Matsuda, F., … Matsuda, F. (2003). Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proceedings of the National Academy of Sciences, 100(19), 10972–10976. https://doi.org/10.1073/pnas.1834399100
Topalglu, A. K., Tello, J. A., Kotan, L. D., Ozbek, M. N., Yilmaz, M. B., Erdogan, S., … Yuksel, B. (2012). Inactivating KISS1 mutation and hypogonadotropic hypogonadism. Obstetrical {\&} Gynecological Survey, 67(6), 352–353. https://doi.org/10.1097/ogx.0b013e31825bc1be
Hiden, U., Bilban, M., Knöfler, M., & Desoye, G. (2007). Kisspeptins and the placenta: Regulation of trophoblast invasion. Reviews in endocrine & metabolic disorders, 8(1), 31–39. https://doi.org/10.1007/s11154-007-9030-8
Padilla, S. L., Perez, J. G., Ben-Hamo, M., Johnson, C. W., Sanchez, R. E. A., Bussi, I. L., … de la Iglesia, H. O. (2019). Kisspeptin neurons in the arcuate nucleus of the hypothalamus orchestrate circadian rhythms and metabolism. Current Biology, 29(4), 592–604. https://doi.org/10.1016/j.cub.2019.01.022
Diamantopoulou, Z., Castro-Giner, F., Schwab, F. D., Foerster, C., Saini, M., Budinjas, S., … Aceto, N. (2022). The metastatic spread of breast cancer accelerates during sleep. Nature, 607(7917), 156–162. https://doi.org/10.1038/s41586-022-04875-y
De Opakua, A. I., Merino, N., Villate, M., Cordeiro, T. N., Ormaza, G., Sánchez-Carbayo, M., … Blanco, F. J. (2017). The metastasis suppressor KISS1 is an intrinsically disordered protein slightly more extended than a random coil. PLoS ONE, 12(2). https://doi.org/10.1371/journal.pone.0172507
Shin, R., Welch, D. R., Mishra, V. K., Nash, K. T., Hurst, D. R., & Rama Krishna, N. (2009). Nuclear magnetic resonance and circular dichroism study of metastin (Kisspeptin-54) structure in solution. Clinical and Experimental Metastasis, 26(6), 527–533. https://doi.org/10.1007/s10585-009-9252-0
Takino, T., Koshikawa, N., Miyamori, H., Tanaka, M., Sasaki, T., Okada, Y., … Sato, H. (2003). Cleavage of metastasis suppressor gene product KiSS-1 protein/metastin by matrix metalloproteinases. Oncogene, 22(30), 4617–4626. https://doi.org/10.1038/sj.onc.1206542
Milton, N. G. N. (2012). In vitro activities of kissorphin, a novel hexapeptide KiSS-1 derivative, in neuronal cells. Journal of Amino Acids, 2012, 1–6. https://doi.org/10.1155/2012/691463
Yeo, R. L., Tsunekawa, K., Mi, J. M., Haet, N. U., Hwang, J. I., Osugi, T., … Tsutsui, K. (2009). Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology, 150(6), 2837–2846. https://doi.org/10.1210/en.2008-1679
Akazome, Y., Kanda, S., Okubo, K., & Oka, Y. (2010). Functional and evolutionary insights into vertebrate kisspeptin systems from studies of fish brain. Journal of Fish Biology, 76(1), 161–182. https://doi.org/10.1111/j.1095-8649.2009.02496.x
Harihar, S., Pounds, K. M., Iwakuma, T., Seidah, N. G., & Welch, D. R. (2014). Furin is the major proprotein convertase required for KISS1-to-Kisspeptin processing. PLoS ONE, 9(1). https://doi.org/10.1371/journal.pone.0084958
Navenot, J.-M., Fujii, N., & Peiper, S. C. (2009). KiSS1 metastasis suppressor gene product induces suppression of tyrosine kinase receptor signaling to Akt, tumor necrosis factor family ligand expression, and apoptosis. Molecular Pharmacology, 75(5), 1074–1083. https://doi.org/10.1124/mol.108.054270
Navenot, J.-M., Fujii, N., & Peiper, S. C. (2009). Activation of Rho and Rho-associated kinase by GPR54 and KiSS1 metastasis suppressor gene product induces changes of cell morphology and contributes to apoptosis. Molecular Pharmacology, 75(6), 1300–1306. https://doi.org/10.1124/mol.109.055095
Cho, S. G., Li, D., Stafford, L. J., Luo, J., Rodriguez-Villanueva, M., Wang, Y., & Liu, M. (2009). KiSS1 suppresses TNFα-induced breast cancer cell invasion via an inhibition of RhoA-mediated NF-κB activation. Journal of Cellular Biochemistry, 107(6), 1139–1149. https://doi.org/10.1002/jcb.22216
Harms, J. F., Welch, D. R., & Miele, M. E. (2003). KISS1 metastasis suppression and emergent pathways. Clinical and Experimental Metastasis, 20, 11–18. https://doi.org/10.1023/A:1022530100931
Teng, Y., Liu, M., & Cowell, J. K. (2011). Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. International Journal of Cancer, 129(12), 2825–2835. https://doi.org/10.1002/ijc.25964
Arab, K., Smith, L. T., Gast, A., Weichenhan, D., Huang, J. P. H., Claus, R., … Plass, C. (2011). Epigenetic deregulation of TCF21 inhibits metastasis suppressor KISS1 in metastatic melanoma. Carcinogenesis, 32(10), 1467–1473. https://doi.org/10.1093/carcin/bgr138
Dotterweich, J., Tower, R. J., Brandl, A., Müller, M., Hofbauer, L. C., Beilhack, A., … Jakob, F. (2016). The KISS1 receptor as an in vivo microenvironment imaging biomarker of multiple myeloma bone disease. PLoS ONE, 11(5). https://doi.org/10.1371/journal.pone.0155087
Lu, X., Mu, E., & Wei, Y. (2011). VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging α4β1-positive osteoclast progenitors. Cancer Cell, 20(6), 701–714. https://doi.org/10.1016/j.ccr.2011.11.002
Barkan, D., Kleinman, H., & Simmons, J. L. (2008). Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Research, 68(15), 6241–6250. https://doi.org/10.1158/0008-5472.CAN-07-6849
Akhtar, M., Haider, A., Rashid, S., & Al-Nabet, A. D. M. H. (2019). Paget’s “seed and Soil” theory of cancer metastasis: An idea whose time has come. Advances in Anatomic Pathology, 26(1), 69–74. https://doi.org/10.1097/PAP.0000000000000219
Lee, K. H., & Kim, J. R. (2009). Kiss-1 suppresses MMP-9 expression by activating p38 MAP kinase in human stomach cancer. Oncology Research, 18(2–3), 107–116. https://doi.org/10.3727/096504009789954591
Kaverina, N., Borovjagin, A. V, Kadagidze, Z., Baryshnikov, A., Baryshnikova, M., Malin, D., … Ulasov, I. V. (2017). Astrocytes promote progression of breast cancer metastases to the brain via a KISS1-mediated autophagy. Autophagy, 13(11), 1905–1923. https://doi.org/10.1080/15548627.2017.1360466
Liu, W., Beck, B. H., Vaidya, K. S., Nash, K. T., Feeley, K. P., Ballinger, S. W., … Welch, D. R. (2014). Metastasis suppressor KISS1 Seems to reverse the warburg effect by enhancing mitochondrial biogenesis. Cancer Research, 74(3), 954–963. https://doi.org/10.1158/0008-5472.CAN-13-1183
Tsuruo, T., Iida, H., Makishima, F., Yamori, T., Kawabata, H., Tsukagoshi, S., & Sakurai, Y. (1985). Inhibition of spontaneous and experimental tumor metastasis by the calcium antagonist verapamil. Cancer Chemotherapy and Pharmacology, 14(1), 30–33. https://doi.org/10.1007/BF00552721
Welch, D. R., Harper, D. E., & Yohem, K. H. (1993). U-77,863: A novel cinnanamide isolated from Streptomyces griseoluteus that inhibits cancer invasion and metastasis. Clinical & experimental metastasis, 11(2), 201–212. https://doi.org/10.1007/BF00114978
Peruzzo, R., Costa, R., Bachmann, M., Leanza, L., & Szabò, I. (2020). Mitochondrial metabolism, contact sites and cellular calcium signaling: Implications for tumorigenesis. Cancers, 12(9), 2574. https://doi.org/10.3390/cancers12092574
McCormack, J. G., & Denton, R. M. (1993). Mitochondrial Ca2+ transport and the role of intramitochondrial Ca2+ in the regulation of energy metabolism. Developmental Neuroscience, 15(3–5), 165–173. https://doi.org/10.1159/000111332
Manley, S. J., Liu, W., & Welch, D. R. (2017). The KISS1 metastasis suppressor appears to reverse the Warburg effect by shifting from glycolysis to mitochondrial beta-oxidation. Journal of Molecular Medicine, 95(9), 951–963. https://doi.org/10.1007/s00109-017-1552-2
Ibrahim-Hashim, A., & Estrella, V. (2019). Acidosis and cancer: From mechanism to neutralization. Cancer and Metastasis Reviews, 38, 149–155. https://doi.org/10.1007/s10555-019-09787-4
Liu, W., Beck, B. H., Vaidya, K. S., Nash, K. T., Feeley, K. P., Ballinger, S. W., … Welch, D. R. (2014). Metastasis suppressor KISS1 seems to reverse the Warburg effect by enhancing mitochondrial biogenesis. Cancer Research, 74(3), 954–963. https://doi.org/10.1158/0008-5472.CAN-13-1183
Pranzini, E., Raugei, G., & Taddei, M. L. (2022). Metabolic features of tumor dormancy: Possible therapeutic strategies. Cancers, 14(3). https://doi.org/10.3390/cancers14030547
Trevisan, C. M., Montagna, E., De Oliveira, R., Christofolini, D. M., Barbosa, C. P., Crandall, K. A., & Bianco, B. (2018). Kisspeptin/GPR54 system: What do we know about its role in human reproduction? Cellular Physiology and Biochemistry, 49, 1259–1276. https://doi.org/10.1159/000493406
De Tassigny, X. D. A., Jayasena, C., Murphy, K. G., Dhillo, W. S., & Colledge, W. H. (2017). Mechanistic insights into the more potent effect of KP-54 compared to KP-10 in vivo. PLoS ONE, 12(5). https://doi.org/10.1371/journal.pone.0176821
Ciaramella, V., Della Corte, C. M., Ciardiello, F., & Morgillo, F. (2018). Kisspeptin and cancer: Molecular interaction, biological functions, and future perspectives. Frontiers in Endocrinology, 9(115). https://doi.org/10.3389/fendo.2018.00115
Werner-Klein, M., & Klein, C. A. (2019). Therapy resistance beyond cellular dormancy. Nature Cell Biology, 21(2), 117–119. https://doi.org/10.1038/s41556-019-0276-7
Sleeman, J., & Steeg, P. S. (2010). Cancer metastasis as a therapeutic target. European journal of cancer (Oxford, England: 1990), 46(7), 1177–1180. https://doi.org/10.1016/j.ejca.2010.02.039
Young, E. D., Strom, K., Tsue, A. F., Usset, J. L., MacPherson, S., McGuire, J. T., & Welch, D. R. (2018). Automated quantitative image analysis for ex vivo metastasis assays reveals differing lung composition requirements for metastasis suppression by KISS1. Clinical and Experimental Metastasis, 35(1–2), 77–86. https://doi.org/10.1007/s10585-018-9882-1
Acknowledgements
Work done in SH’s lab has been generously funded by a grant from Indian Council of Medical Research (ICMR-2021-8665-F1). Work done in DRW’s lab has been generously funded by grants from the U.S. National Cancer Institute (RO1-CA62168; P30-CA168524), the National Foundation for Cancer Research, METAvivor Research and Services Inc., and Theresa’s Research Foundation. We apologize to any authors whose work was omitted due to article guidelines.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Harihar, S., Welch, D.R. KISS1 metastasis suppressor in tumor dormancy: a potential therapeutic target for metastatic cancers?. Cancer Metastasis Rev 42, 183–196 (2023). https://doi.org/10.1007/s10555-023-10090-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-023-10090-6