Abstract
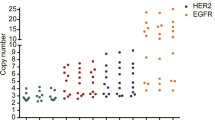
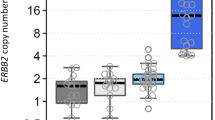
Gene amplification is an important factor for altered gene expression in breast cancers. TOP2A-amplification often occurs concomitantly with HER2 amplification, and it has been suggested to be predictive for the response to anthracycline chemotherapy. This study assessed the correlation between HER2 status and TOP2A co-amplification, the possible association of TOP2A single-nucleotide polymorphisms with the frequency of this co-amplification as well as confirmation of association with outcome. HER2 and TOP2A amplification were analyzed in a tissue microarray from a clinical cohort study. Additionally, a common genetic variant (rs13695) in the TOP2A gene was genotyped in germline DNA. HER2 gene amplification was compared with HER2-IHC findings assessed during clinical routine work, and the association between all the biomarkers analyzed and the clinical outcome was determined. As an exploratory aim, rs13695 genotypes were compared with TOP2A amplification status. HER2 amplification was seen in 101 of 628 (16.1 %) and TOP2A amplification in 32 (5.1 %) cancers. No TOP2A amplification occurred without HER2 co-amplification. HER2 amplification was found in 8, 13.6, and 55.1 % of patients with HER2-IHC 0/1+, 2+, and 3+ tumors, respectively. HER2-IHC was not associated with an effect on the prognosis, but HER2-FISH was. There was an association between the rs13695 genotype and TOP2A amplification status (P = 0.03). Although there was a significant correlation between HER2 status determined by IHC and HER2 by FISH, only HER2 gene amplification status by FISH was correlated with outcome indicating greater utility for FISH in routine clinical settings.


Similar content being viewed by others
References
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352. doi:10.1038/nature10983
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 235(4785):177–182
Press MF, Sauter G, Buyse M, Bernstein L, Guzman R, Santiago A, Villalobos IE, Eiermann W, Pienkowski T, Martin M, Robert N, Crown J, Bee V, Taupin H, Flom KJ, Tabah-Fisch I, Pauletti G, Lindsay MA, Riva A, Slamon DJ (2011) Alteration of topoisomerase II-alpha gene in human breast cancer: association with responsiveness to anthracycline-based chemotherapy. J Clin Oncol 29(7):859–867. doi:10.1200/JCO.2009.27.5644
Di Leo A, Desmedt C, Bartlett JM, Piette F, Ejlertsen B, Pritchard KI, Larsimont D, Poole C, Isola J, Earl H, Mouridsen H, O’Malley FP, Cardoso F, Tanner M, Munro A, Twelves CJ, Sotiriou C, Shepherd L, Cameron D, Piccart MJ, Buyse M (2011) HER2 and TOP2A as predictive markers for anthracycline-containing chemotherapy regimens as adjuvant treatment of breast cancer: a meta-analysis of individual patient data. Lancet Oncol 12(12):1134–1142. doi:10.1016/S1470-2045(11)70231-5
Thor AD, Berry DA, Budman DR, Muss HB, Kute T, Henderson IC, Barcos M, Cirrincione C, Edgerton S, Allred C, Norton L, Liu ET (1998) erbB-2, p53, and efficacy of adjuvant therapy in lymph node-positive breast cancer. J Natl Cancer Inst 90(18):1346–1360
Pritchard KI, Shepherd LE, O’Malley FP, Andrulis IL, Tu D, Bramwell VH, Levine MN (2006) HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med 354(20):2103–2111. doi:10.1056/NEJMoa054504
Piccart-Gebhart MJ (2006) Anthracyclines and the tailoring of treatment for early breast cancer. N Engl J Med 354(20):2177–2179. doi:10.1056/NEJMe068065
Paik S, Bryant J, Park C, Fisher B, Tan-Chiu E, Hyams D, Fisher ER, Lippman ME, Wickerham DL, Wolmark N (1998) erbB-2 and response to doxorubicin in patients with axillary lymph node-positive, hormone receptor-negative breast cancer. J Natl Cancer Inst 90(18):1361–1370
Muss HB, Thor AD, Berry DA, Kute T, Liu ET, Koerner F, Cirrincione CT, Budman DR, Wood WC, Barcos M et al (1994) c-erbB-2 expression and response to adjuvant therapy in women with node-positive early breast cancer. N Engl J Med 330(18):1260–1266. doi:10.1056/NEJM199405053301802
Luftner D, Lux MP, Maass N, Schutz F, Schwidde I, Fasching PA, Fehm T, Janni W, Kummel S, Kolberg HC (2012) Advances in breast cancer: looking back over the year. Geburtsh Frauenheilk 72(12):1117–1129. doi:10.1055/s-0032-1328084
Kolberg HC, Luftner D, Lux MP, Maass N, Schutz F, Fasching PA, Fehm T, Janni W, Kummel S (2012) Breast cancer 2012-new aspects. Geburtsh Frauenheilk 72(7):602–615. doi:10.1055/s-0032-1315131
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF, American Society of Clinical O, College of American P (2013) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 31(31):3997–4013. doi:10.1200/JCO.2013.50.9984
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25(1):118–145. doi:10.1200/JCO.2006.09.2775
Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF (2009) Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol 27(8):1323–1333. doi:10.1200/JCO.2007.14.8197
van de Vijver M, van de Bersselaar R, Devilee P, Cornelisse C, Peterse J, Nusse R (1987) Amplification of the neu (c-erbB-2) oncogene in human mammmary tumors is relatively frequent and is often accompanied by amplification of the linked c-erbA oncogene. Mol Cell Biol 7(5):2019–2023
Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC (1995) Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 28(3):367–376. doi:10.1006/geno.1995.1163
Smith K, Houlbrook S, Greenall M, Carmichael J, Harris AL (1993) Topoisomerase II alpha co-amplification with erbB2 in human primary breast cancer and breast cancer cell lines: relationship to m-AMSA and mitoxantrone sensitivity. Oncogene 8(4):933–938
Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R (1996) Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res 56(17):3886–3890
Konecny GE, Pauletti G, Untch M, Wang HJ, Mobus V, Kuhn W, Thomssen C, Harbeck N, Wang L, Apple S, Janicke F, Slamon DJ (2010) Association between HER2, TOP2A, and response to anthracycline-based preoperative chemotherapy in high-risk primary breast cancer. Breast Cancer Res Treat 120(2):481–489. doi:10.1007/s10549-010-0744-z
Salmen J, Rack B, Ortmann U, Sommer H, Beckmann MW, Lichtenegger W, Janni W, Schneeweiss A (2011) Simultaneous study of docetaxel based anthracycline free adjuvant treatment evaluation, as well as life style intervention strategies: the German SUCCESS C trial. Breast 20:S66–S66
Baldwin RM, Owzar K, Zembutsu H, Chhibber A, Kubo M, Jiang C, Watson D, Eclov RJ, Mefford J, McLeod HL, Friedman PN, Hudis CA, Winer EP, Jorgenson EM, Witte JS, Shulman LN, Nakamura Y, Ratain MJ, Kroetz DL (2012) A genome-wide association study identifies novel loci for paclitaxel-induced sensory peripheral neuropathy in CALGB 40101. Clin Cancer Res 18(18):5099–5109. doi:10.1158/1078-0432.CCR-12-1590
Cheang M, Chia SK, Tu D, Jiang S, LE Shepherd, Pritchard KI, Nielsen TO (2009) Anthracyclines in basal breast cancer: the NCIC-CTG trial MA5 comparing adjuvant CMF to CEF. J Clin Oncol 27(15S):a519
Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, Geiler S, Ringleff K, Oeser S, Weihbrecht S, Schulz-Wendtland R, Hartmann A, Beckmann MW, Strick R (2008) Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res Treat 112(1):89–98. doi:10.1007/s10549-007-9822-2
Heusinger K, Loehberg CR, Haeberle L, Jud SM, Klingsiek P, Hein A, Bayer CM, Rauh C, Uder M, Cavallaro A, May MS, Adamietz B, Schulz-Wendtland R, Wittenberg T, Wagner F, Beckmann MW, Fasching PA (2011) Mammographic density as a risk factor for breast cancer in a German case-control study. Eur J Cancer Prev 20(1):1–8. doi:10.1097/CEJ.0b013e328341e2ce
Schrauder M, Frank S, Strissel PL, Lux MP, Bani MR, Rauh C, Sieber CC, Heusinger K, Hartmann A, Schulz-Wendtland R, Strick R, Beckmann MW, Fasching PA (2008) Single nucleotide polymorphism D1853N of the ATM gene may alter the risk for breast cancer. J Cancer Res Clin Oncol 134(8):873–882. doi:10.1007/s00432-008-0355-9
Fasching PA, Ekici AB, Wachter DL, Hein A, Bayer CM, Haberle L, Loehberg C, Schneider M, Jud SM, Heusinger K, Rubner M, Rauh C, bani M, Lux MP, Schulz-Wendtland R, Hartmann A, Beckmann MW (2013) Breast cancer risk: from genetics to molecular understanding of pathogenesis. Geburtsh Frauenheilk 73:1228–1235. doi:10.1055/s-0033-1360178
Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, Bojesen SE, Nordestgaard BG, Flyger H, Milne RL, Arias JI, Menendez P, Benitez J, Chang-Claude J, Hein R, Wang-Gohrke S, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Margolin S, Mannermaa A, Kosma VM, Kataja V, Beesley J, Chen X, Chenevix-Trench G, Couch FJ, Olson JE, Fredericksen ZS, Wang X, Giles GG, Severi G, Baglietto L, Southey MC, Devilee P, Tollenaar RA, Seynaeve C, Garcia-Closas M, Lissowska J, Sherman ME, Bolton KL, Hall P, Czene K, Cox A, Brock IW, Elliott GC, Reed MW, Greenberg D, Anton-Culver H, Ziogas A, Humphreys M, Easton DF, Caporaso NE, Pharoah PD (2010) Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. J Natl Cancer Inst 102(9):650–662. doi:10.1093/jnci/djq057
Baglietto L, Hooning MJ, Martens JW, Jager A, Kriege M, Lindblom A, Margolin S, Couch FJ, Stevens KN, Olson JE, Kosel M, Cross SS, Balasubramanian SP, Reed MW, Miron A, John EM, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Kauppila S, Burwinkel B, Marme F, Schneeweiss A, Sohn C, Chenevix-Trench G, Lambrechts D, Dieudonne AS, Hatse S, van Limbergen E, Benitez J, Milne RL, Zamora MP, Perez JI, Bonanni B, Peissel B, Loris B, Peterlongo P, Rajaraman P, Schonfeld SJ, Anton-Culver H, Devilee P, Beckmann MW, Slamon DJ, Phillips KA, Figueroa JD, Humphreys MK, Easton DF, Schmidt MK (2012) The role of genetic breast cancer susceptibility variants as prognostic factors. Hum Mol Genet 21(17):3926–3939. doi:10.1093/hmg/dds159
Lambrechts D, Truong T, Justenhoven C, Humphreys MK, Wang J, Hopper JL, Dite GS, Apicella C, Southey MC, Schmidt MK, Broeks A, Cornelissen S, Hien RV, Sawyer E, Tomlinson I, Kerin M, Miller N, Milne RL, Zamora MP, Perez JI, Benitez J, Hamann U, Ko YD, Bruning T, Chang-Claude J, Eilber U, Hein R, Nickels S, Flesch-Janys D, Wang-Gohrke S, John EM, Miron A, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Chenevix-Trench G, Beesley J, Chen X, Menegaux F, Cordina-Duverger E, Shen CY, Yu JC, Wu PE, Hou MF, Andrulis IL, Selander T, Glendon G, Mulligan AM, Anton-Culver H, Ziogas A, Muir KR, Lophatananon A, Rattanamongkongul S, Puttawibul P, Jones M, Orr N, Ashworth A, Swerdlow A, Severi G, Baglietto L, Giles G, Southey M, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Yesilyurt BT, Neven P, Paridaens R, Wildiers H, Brenner H, Muller H, Arndt V, Stegmaier C, Meindl A, Schott S, Bartram CR, Schmutzler RK, Cox A, Brock IW, Elliott G, Cross SS, Fasching PA, Schulz-Wendtland R, Ekici AB, Beckmann MW, Fletcher O, Johnson N, Dos Santos Silva I, Peto J, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Dork T, Schurmann P, Bremer M, Hillemanns P, Bogdanova NV, Antonenkova NN, Rogov YI, Karstens JH, Khusnutdinova E, Bermisheva M, Prokofieva D, Gancev S, Jakubowska A, Lubinski J, Jaworska K, Durda K, Nordestgaard BG, Bojesen SE, Lanng C, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Radice P, Peterlongo P, Manoukian S, Bernard L, Couch FJ, Olson JE, Wang X, Fredericksen Z, Alnsss GG, Kristensen V, Borresen-Dale AL, Devilee P, Tollenaar RA, Seynaeve CM, Hooning MJ, Garcia-Closas M, Chanock SJ, Lissowska J, Sherman ME, Hall P, Liu J, Czene K, Kang D, Yoo KY, Noh DY, Lindblom A, Margolin S, Dunning AM, Pharoah PD, Easton DF, Guenel P, Brauch H (2012) 11q13 is a susceptibility locus for hormone receptor positive breast cancer. Hum Mutat. doi:10.1002/humu.22089
Hein R, Maranian M, Hopper JL, Kapuscinski MK, Southey MC, Park DJ, Schmidt MK, Broeks A, Hogervorst FB, Bueno-de-Mesquit HB, Muir KR, Lophatananon A, Rattanamongkongul S, Puttawibul P, Fasching PA, Hein A, Ekici AB, Beckmann MW, Fletcher O, Johnson N, Dos Santos Silva I, Peto J, Sawyer E, Tomlinson I, Kerin M, Miller N, Marmee F, Schneeweiss A, Sohn C, Burwinkel B, Guenel P, Cordina-Duverger E, Menegaux F, Truong T, Bojesen SE, Nordestgaard BG, Flyger H, Milne RL, Perez JI, Zamora MP, Benitez J, Anton-Culver H, Ziogas A, Bernstein L, Clarke CA, Brenner H, Muller H, Arndt V, Stegmaier C, Rahman N, Seal S, Turnbull C, Renwick A, Meindl A, Schott S, Bartram CR, Schmutzler RK, Brauch H, Hamann U, Ko YD, Wang-Gohrke S, Dork T, Schurmann P, Karstens JH, Hillemanns P, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Bogdanova NV, Zalutsky IV, Antonenkova NN, Bermisheva M, Prokovieva D, Farahtdinova A, Khusnutdinova E, Lindblom A, Margolin S, Mannermaa A, Kataja V, Kosma VM, Hartikainen J, Chen X, Beesley J, Investigators K, Lambrechts D, Zhao H, Neven P, Wildiers H, Nickels S, Flesch-Janys D, Radice P, Peterlongo P, Manoukian S, Barile M, Couch FJ, Olson JE, Wang X, Fredericksen Z, Giles GG, Baglietto L, McLean CA, Severi G, Offit K, Robson M, Gaudet MM, Vijai J, Alnaes GG, Kristensen V, Borresen-Dale AL, John EM, Miron A, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Figueroa JD, Garcia-Closas M, Lissowska J, Sherman ME, Hooning M, Martens JW, Seynaeve C, Collee M, Hall P, Humpreys K, Czene K, Liu J, Cox A, Brock IW, Cross SS, Reed MW, Ahmed S, Ghoussaini M, Pharoah PD, Kang D, Yoo KY, Noh DY, Jakubowska A, Jaworska K, Durda K, Zlowocka E, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Shen CY, Yu JC, Hsu HM, Hou MF, Orr N, Schoemaker M, Ashworth A, Swerdlow A, Trentham-Dietz A, Newcomb PA, Titus L, Egan KM, Chenevix-Trench G, Antoniou AC, Humphreys MK, Morrison J, Chang-Claude J, Easton DF, Dunning AM (2012) Comparison of 6q25 breast cancer hits from Asian and European Genome Wide Association Studies in the Breast Cancer Association Consortium (BCAC). PLoS One 7(8):e42380. doi:10.1371/journal.pone.0042380
Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, Michailidou K, Stram DO, Beckmann L, Rhie SK, Ambrosone CB, Aittomaki K, Amiano P, Apicella C, Baglietto L, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Brauch H, Brinton L, Bui QM, Buring JE, Buys SS, Campa D, Carpenter JE, Chasman DI, Chang-Claude J, Chen C, Clavel-Chapelon F, Cox A, Cross SS, Czene K, Deming SL, Diasio RB, Diver WR, Dunning AM, Durcan L, Ekici AB, Fasching PA, Fejerman L, Figueroa JD, Fletcher O, Flesch-Janys D, Gaudet MM, Gerty SM, Rodriguez-Gil JL, Giles GG, van Gils CH, Godwin AK, Graham N, Greco D, Hall P, Hankinson SE, Hartmann A, Hein R, Heinz J, Hoover RN, Hopper JL, Hu JJ, Huntsman S, Ingles SA, Irwanto A, Isaacs C, Jacobs KB, John EM, Justenhoven C, Kaaks R, Kolonel LN, Coetzee GA, Lathrop M, Le Marchand L, Lee AM, Lee IM, Lesnick T, Lichtner P, Liu J, Lund E, Makalic E, Martin NG, McLean CA, Meijers-Heijboer H, Meindl A, Miron P, Monroe KR, Montgomery GW, Muller-Myhsok B, Nickels S, Nyante SJ, Olswold C, Overvad K, Palli D, Park DJ, Palmer JR, Pathak H, Peto J, Pharoah P, Rahman N, Rivadeneira F, Schmidt DF, Schmutzler RK, Slager S, Southey MC, Stevens KN, Sinn HP, Press MF, Ross E, Riboli E, Ridker PM, Schumacher FR, Severi G, Silva ID, Stone J, Sund M, Tapper WJ, Thun MJ, Travis RC, Turnbull C, Uitterlinden AG, Waisfisz Q, Wang X, Wang Z, Weaver J, Schulz-Wendtland R, Wilkens LR, Van Den Berg D, Zheng W, Ziegler RG, Ziv E, Nevanlinna H, Easton DF, Hunter DJ, Henderson BE, Chanock SJ, Garcia-Closas M, Kraft P, Haiman CA, Vachon CM (2012) A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. doi:10.1093/hmg/dds381
Beckmann MW, Brucker C, Hanf V, Rauh C, Bani MR, Knob S, Petsch S, Schick S, Fasching PA, Hartmann A, Lux MP, Haberle L (2011) Quality assured health care in certified breast centers and improvement of the prognosis of breast cancer patients. Onkologie 34(7):362–367. doi:10.1159/000329601
Mass RD, Press MF, Anderson S, Cobleigh MA, Vogel CL, Dybdal N, Leiberman G, Slamon DJ (2005) Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer 6(3):240–246. doi:10.3816/CBC.2005.n.026
Press MF, Sauter G, Bernstein L, Villalobos IE, Mirlacher M, Zhou JY, Wardeh R, Li YT, Guzman R, Ma Y, Sullivan-Halley J, Santiago A, Park JM, Riva A, Slamon DJ (2005) Diagnostic evaluation of HER-2 as a molecular target: an assessment of accuracy and reproducibility of laboratory testing in large, prospective, randomized clinical trials. Clinical Cancer Res 11(18):6598–6607. doi:10.1158/1078-0432.CCR-05-0636
Gown AM, Goldstein LC, Barry TS, Kussick SJ, Kandalaft PL, Kim PM, Tse CC (2008) High concordance between immunohistochemistry and fluorescence in situ hybridization testing for HER2 status in breast cancer requires a normalized IHC scoring system. Mod Pathol 21(10):1271–1277. doi:10.1038/modpathol.2008.83
Yaziji H, Goldstein LC, Barry TS, Werling R, Hwang H, Ellis GK, Gralow JR, Livingston RB, Gown AM (2004) HER-2 testing in breast cancer using parallel tissue-based methods. J Am Med Assoc 291(16):1972–1977. doi:10.1001/jama.291.16.1972
Grambsch PM, Therneau TM (1994) Proportional hazards tests and diagnostics based on weighted residuals. Biometrika 81(3):515–526
Dekker TJ, Borg ST, Hooijer GK, Meijer SL, Wesseling J, Boers JE, Schuuring E, Bart J, van Gorp J, Mesker WE, Kroep JR, Smit VT, van de Vijver MJ (2012) Determining sensitivity and specificity of HER2 testing in breast cancer using a tissue micro-array approach. Breast Cancer Res 14(3):R93. doi:10.1186/bcr3208
Dybdal N, Leiberman G, Anderson S, McCune B, Bajamonde A, Cohen RL, Mass RD, Sanders C, Press MF (2005) Determination of HER2 gene amplification by fluorescence in situ hybridization and concordance with the clinical trials immunohistochemical assay in women with metastatic breast cancer evaluated for treatment with trastuzumab. Breast Cancer Res Treat 93(1):3–11. doi:10.1007/s10549-004-6275-8
Pauletti G, Dandekar S, Rong H, Ramos L, Peng H, Seshadri R, Slamon DJ (2000) Assessment of methods for tissue-based detection of the HER-2/neu alteration in human breast cancer: a direct comparison of fluorescence in situ hybridization and immunohistochemistry. J Clin Oncol 18(21):3651–3664
Ciampa A, Xu B, Ayata G, Baiyee D, Wallace J, Wertheimer M, Edmiston K, Khan A (2006) HER-2 status in breast cancer: correlation of gene amplification by FISH with immunohistochemistry expression using advanced cellular imaging system. Appl Immunohistochem Mol Morphol 14(2):132–137. doi:10.1097/01.pai.0000150516.75567.13
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ (1999) Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER-2/neu in breast cancer. J Clin Oncol 17(7):1974–1982
Owens MA, Horten BC, Da Silva MM (2004) HER2 amplification ratios by fluorescence in situ hybridization and correlation with immunohistochemistry in a cohort of 6556 breast cancer tissues. Clin Breast Cancer 5(1):63–69
Perez EA, Suman VJ, Davidson NE, Martino S, Kaufman PA, Lingle WL, Flynn PJ, Ingle JN, Visscher D, Jenkins RB (2006) HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol 24(19):3032–3038. doi:10.1200/JCO.2005.03.4744
Bartlett JM, Going JJ, Mallon EA, Watters AD, Reeves JR, Stanton P, Richmond J, Donald B, Ferrier R, Cooke TG (2001) Evaluating HER2 amplification and overexpression in breast cancer. J Pathol 195(4):422–428. doi:10.1002/path.971
Press MF, Hung G, Godolphin W, Slamon DJ (1994) Sensitivity of HER-2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 54(10):2771–2777
Dowsett M, Bartlett J, Ellis IO, Salter J, Hills M, Mallon E, Watters AD, Cooke T, Paish C, Wencyk PM, Pinder SE (2003) Correlation between immunohistochemistry (HercepTest) and fluorescence in situ hybridization (FISH) for HER-2 in 426 breast carcinomas from 37 centres. J Pathol 199(4):418–423. doi:10.1002/path.1313
Lal P, Salazar PA, Hudis CA, Ladanyi M, Chen B (2004) HER-2 testing in breast cancer using immunohistochemical analysis and fluorescence in situ hybridization: a single-institution experience of 2,279 cases and comparison of dual-color and single-color scoring. Am J Clin Pathol 121(5):631–636. doi:10.1309/VE78-62V2-646B-R6EX
Al-Khattabi H, Kelany A, Buhmeida A, Al-Maghrabi J, Lari S, Chaudhary A, Gari M, Abuzenadah A, Al-Qahtani M (2010) Evaluation of HER-2/neu gene amplification by fluorescence in situ hybridization and immunohistochemistry in Saudi female breast cancer. Anticancer Res 30(10):4081–4088
Kovacs A, Stenman G (2010) HER2-testing in 538 consecutive breast cancer cases using FISH and immunohistochemistry. Pathol Res Pract 206(1):39–42. doi:10.1016/j.prp.2009.08.003
Meijer SL, Wesseling J, Smit VT, Nederlof PM, Hooijer GK, Ruijter H, Arends JW, Kliffen M, van Gorp JM, Sterk L, van de Vijver MJ (2011) HER2 gene amplification in patients with breast cancer with equivocal IHC results. J Clin Pathol 64(12):1069–1072. doi:10.1136/jclinpath-2011-200019
Bilous M, Morey AL, Armes JE, Bell R, Button PH, Cummings MC, Fox SB, Francis GD, Waite B, McCue G, Raymond WA, Robbins PD, Farshid G (2012) Assessing HER2 amplification in breast cancer: findings from the Australian in situ Hybridization Program. Breast Cancer Res Treat 134(2):617–624. doi:10.1007/s10549-012-2093-6
Tvrdik D, Stanek L, Skalova H, Dundr P, Velenska Z, Povysil C (2012) Comparison of the IHC, FISH, SISH and qPCR methods for the molecular diagnosis of breast cancer. Mol Med Rep 6(2):439–443. doi:10.3892/mmr.2012.919
Yang YL, Fan Y, Lang RG, Gu F, Ren MJ, Zhang XM, Yin D, Fu L (2012) Genetic heterogeneity of HER2 in breast cancer: impact on HER2 testing and its clinicopathologic significance. Breast Cancer Res Treat 134(3):1095–1102. doi:10.1007/s10549-012-2046-0
Bohn OL, Sanchez-Sosa S, Ibarra JA, Rogers LW (2011) Fixation time and HER2/neu assessment. Am J Clin Pathol 135(6):979–980. doi:10.1309/AJCPVRQS9UQTCZE5
Goldsmith JD, Allred DC, Beasley MB, Eisen R, Fulton RS, Gown AM, Hammond ME (2011) Fixation time does not affect expression of HER2/neu. Am J Clin Pathol 135(3):484. doi:10.1309/AJCPO1ZG6FERBGZB
Ibarra JA, Rogers LW (2010) Fixation time does not affect expression of HER2/neu: a pilot study. Am J Clin Pathol 134(4):594–596. doi:10.1309/AJCPAIJPSN4A9MJI
Jacobs TW, Gown AM, Yaziji H, Barnes MJ, Schnitt SJ (1999) Specificity of HercepTest in determining HER-2/neu status of breast cancers using the United States Food and Drug Administration-approved scoring system. J Clin Oncol 17(7):1983–1987
Gown AM, Goldstein LC (2011) The knowns and the unknowns in HER2 testing in breast cancer. Am J Clin Pathol 136(1):5–6. doi:10.1309/AJCP53KBPMDRTYDK
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M (2010) The landscape of somatic copy-number alteration across human cancers. Nature 463(7283):899–905. doi:10.1038/nature08822
Fudenberg G, Getz G, Meyerson M, Mirny LA (2011) High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol 29(12):1109–1113. doi:10.1038/nbt.2049
Smith AJ, Zheng D, Palmen J, Pang DX, Woo P, Humphries SE (2012) Effects of genetic variation on chromatin structure and the transcriptional machinery: analysis of the IL6 gene locus. Genes Immun. doi:10.1038/gene.2012.32
Acknowledgments
Research for the present study was supported by a grant from the Bavaria California Technology Center (www.bacatec.de) and grants from the California Breast Cancer Research Program (BCRP 12IB-0155 to MFPress) and Breast Cancer Research Foundation (MFPress). Peter A. Fasching was funded by a grant from the Dr. Mildred Scheel Foundation of German Cancer Aid (Deutsche Krebshilfe e.V.).
Conflict of interest
The authors declare they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fasching, P.A., Weihbrecht, S., Haeberle, L. et al. HER2 and TOP2A amplification in a hospital-based cohort of breast cancer patients: associations with patient and tumor characteristics. Breast Cancer Res Treat 145, 193–203 (2014). https://doi.org/10.1007/s10549-014-2922-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-2922-x