Abstract
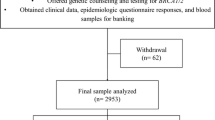
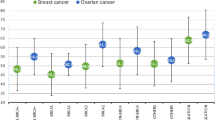
BRCA1/2 mutation status is of paramount importance to identify families at risk of Hereditary Breast and Ovarian Cancer (HBOC). Most HBOC and BRCA1/2 mutation studies have focused on highly selected sub-populations, and few data are available for large population cohorts. For this reason, as part of a regional cancer prevention strategy in North-Central Italy, we set up a population-based screening programme to identify all resident HBOC families, and to determine their BRCA1/2 mutation status. To date, 44 different BRCA1/2 variants have been identified in 55 HBOC families. Of the seven newly reported mutations, only BRCA1 Q284X is clearly deleterious. The analysis of clinical disease characteristics in relation to age of disease onset and family history showed a difference between BRCA1/2 wild type and mutation carrier families. Interestingly, BRCA1/2 mutations were significantly more common in women who developed breast cancer ≤40 years of age than in BRCA1/2 wild type women (50% vs. 29%, respectively, P = 0.005). The family history selection criteria most likely to indicate the presence of deleterious BRCA1/2 mutations are breast cancer ≤35 years (P = 0.012), two first-degree relatives with breast cancer ≤50 years (P = 0.022), and male breast cancer (P = 0.047). The penetrance of BRCA1/2 alterations in our cohort seems to be aligned with other published results. However, new data interpretations have emerged in relation to the clinical criteria and the presence of deleterious mutations. This information shows that a correct and accurate clinical selection could avoid unnecessary molecular tests and could better address genetic analysis and clinical management.

Similar content being viewed by others
References
Easton DF, Pooley KA, Dunning AM et al (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447:1087–1093
Stacey SN, Manolescu A, Sulem P et al (2007) Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 39:865–869
Walsh T, King MC (2007) Ten genes for inherited breast cancer. Cancer Cell 11:103–105
King MC, Marks JH, Mandell JB, et al, New York Breast Cancer Study Group (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 302:643–646
Casadei S, Falcini F, Naldoni C et al (2002) Population-based screening for hereditary breast cancer in a region of North-Central Italy. Int J Mol Med 10:299–305
Cipollini G, Tommasi S, Paradiso A et al (2004) Genetic alterations in hereditary breast cancer. Ann Oncol 15(Suppl 1):I7–I13
Musolino A, Michiara M, Bella MA et al (2005) Molecular profile and clinical variables in BRCA1-positive breast cancers. A population-based study. Tumori 9:505–512
Palomba G, Pisano M, Cossu A et al (2005) Spectrum and prevalence of BRCA1 and BRCA2 germline mutations in Sardinian patients with breast carcinoma through hospital-based screening. Cancer 104:1172–1179
Agata S, Dalla Palma M, Callegaro M et al (2005) Large genomic deletions inactivate the BRCA2 gene in breast cancer families. J Med Genet 42:e64
Kawahara M, Sakayori M, Shiraishi K et al (2004) Identification and evaluation of 55 genetic variations in the BRCA1 and the BRCA2 genes of patients from 50 Japanese breast cancer families. J Hum Genet 49:391–395
Brzovic PS, Meza J, King MC et al (1998) The cancer-predisposing mutation C61G disrupts homodimer formation in the NH2-terminal BRCA1 RING finger domain. J Biol Chem 273:7795–7799
Hashizume R, Fukuda M, Maeda I et al (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276:14537–14540
Ruffner H, Joazeiro CA, Hemmati D et al (2001) Cancer-predisposing mutations within the RING domain of BRCA1: loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc Natl Acad Sci USA 98:5134–5139
Abkevich V, Zharkikh A, Deffenbaugh AM et al (2004) Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet 41:492–507
Orelli BJ, Logsdon JMJ Jr, Bishop DK (2001) Nine novel conserved motifs in BRCA1 identified by the chicken orthologue. Oncogene 20:4433–4438
Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174
Roa BB, Boyd AA, Volcik K et al (1996) Ashkenanzi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet 14:185–187
Antoniou AC, Pharoah PD, Narod S et al (2005) Breast and ovarian cancer risks to carriers of the BRCA1 5382insC and 185delAG and BRCA2 6174delT mutations: a combined analysis of 22 population based studies. J Med Genet 42:602–603
Tommasi S, Crapolicchio A, Lacalamita R et al (2005) BRCA1 mutations and polymorphisms in a hospital-based consecutive series of breast cancer patients from Apulia, Italy. Mutat Res 578:395–405
Goldgar DE, Easton DF, Deffenbaugh AM et al, Breast Cancer Information Core (BIC) Steering Committee (2004) Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet 75:535–544
Hogervorst FB, Nederlof PM, Gille JJ et al (2003) Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res 63:1449–1453
Montagna M, Dalla Palma M, Menin C et al (2003) Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet 12:1055–1061
Hartmann C, John AL, Klaes R et al (2004) Large BRCA1 gene deletions are found in 3% of German high-risk breast cancer families. Hum Mutat 24:534
Agata S, Viel A, Della Puppa L et al (2006) Prevalence of BRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectable BRCA1 and BRCA2 point mutations. Genes Chromosomes Cancer 45:791–797
Walsh T, Casadei S, Coats KH et al (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295:1379–1388
Falchetti M, Lupi R, Rizzolo P et al (2007) BRCA1/BRCA2 rearrangements and CHEK2 common mutations are infrequent in Italian male breast cancer cases. Breast Cancer Res Treat (in press). doi:10.1007/s10549-006-9303-2
Acknowledgements
The Authors wish to thank Prof. Rosella Silvestrini for her invaluable scientific contribution. Thanks also to E. Lucchi for assistance with sample collection.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seymour, I.J., Casadei, S., Zampiga, V. et al. Results of a population-based screening for hereditary breast cancer in a region of North-Central Italy: contribution of BRCA1/2 germ-line mutations. Breast Cancer Res Treat 112, 343–349 (2008). https://doi.org/10.1007/s10549-007-9846-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-007-9846-7