Abstract
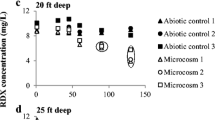
Groundwater contamination by the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is a global problem. Israel’s coastal aquifer was contaminated with RDX. This aquifer is mostly aerobic and we therefore sought aerobic bacteria that might be involved in natural attenuation of the compound in the aquifer. RDX-degrading bacteria were captured by passively sampling the indigenous bacteria onto sterile sediments placed within sampling boreholes. Aerobic RDX biodegradation potential was detected in the sediments sampled from different locations along the plume. RDX degradation with the native sampled consortium was accompanied by 4-nitro-2,4-diazabutanal formation. Two bacterial strains of the genus Rhodococcus were isolated from the sediments and identified as aerobic RDX degraders. The xplA gene encoding the cytochrome P450 enzyme was partially (~500 bp) sequenced from both isolates. The obtained DNA sequences had 99% identity with corresponding gene fragments of previously isolated RDX-degrading Rhodococcus strains. RDX degradation by both strains was prevented by 200 μM of the cytochrome P450 inhibitor metyrapone, suggesting that cytochrome P450 indeed mediates the initial step in RDX degradation. RDX biodegradation activity by the T7 isolate was inhibited in the presence of nitrate or ammonium concentrations above 1.6 and 5.5 mM, respectively (100 mg l−1) while the T9N isolate’s activity was retarded only by ammonium concentrations above 5.5 mM. This study shows that bacteria from the genus Rhodococcus, potentially degrade RDX in the saturated zone as well, following the same aerobic degradation pathway defined for other Rhodococcus species. RDX-degrading activity by the Rhodococcus species isolate T9N may have important implications for the bioremediation of nitrate-rich RDX-contaminated aquifers.



Similar content being viewed by others
References
Alfreider A, Krössbacher M, Psenner R (1997) Groundwater samples do not reflect bacterial densities and activity in subsurface systems. Water Res 31:832–840
Altschul SF, Gish WM, Webb ME, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bej AK, Saul D, Aislabie J (2000) Cold-tolerant alkane-degrading Rhodococcus species from Antarctica. Polar Biol 23:100–105
Bell KS, Philp JC, Aw DWJ, Christofi N (1998) The genus Rhodococcus. J Appl Microbiol 85:195–210
Beller HR (2002) Anaeroic biotransformation of RDX (hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine) by aquifer bacteria using hydrogen as the sole electron donor. Water Res 36:2533–2540
Beller HR, Tiemeier K (2002) Use of liquid chromatography/tandem mass spectrometry to detect distinctive indicators of in situ RDX transformation in contaminated groundwater. Environ Sci Technol 36:2060–2066
Bernstein A, Ronen Z, Adar E, Nativ R, Lowag H, Stichler W, Meckenstock RU (2008) Compound-specific isotope analysis of RDX and stable isotope fractionation during aerobic and anaerobic biodegradation. Environ Sci Technol 42:7772–7777
Bernstein A, Adar E, Ronen Z, Lowag H, Stichler W, Meckenstock RU (2010) Quantifying RDX biodegradation in groundwater using δ15N isotope analysis. J Contam Hydrol 111:25–35
Bhushan B, Trott S, Spain JC, Halasz A, Paquet L, Hawari J (2003) Biotransformation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) by a rabbit liver cytochrome P450: insight into the mechanism of RDX biodegradation by Rhodococcus sp. strain DN22. Appl Environ Microbiol 69:1347–1351
Bradley P, Dinicola R (2005) RDX (hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine) biodegradation in aquifer sediments under manganese-reducing conditions. Bioremediation 9:1–8
Brenner A, Ronen Z, Harel Y, Abeliovich A (2000) Degradation of RDX during biological treatment of munitions waste. Water Environ Res 72:469–475
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145
Coleman NV, Nelson DR, Duxbury T (1998) Aerobic biodegradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol Biochem 30:1159–1167
Coleman NV, Spain JC, Duxbury T (2002) Evidence that RDX biodegradation by Rhodococcus strain DN22 is plasmid-borne and involves a cytochrome P450. J Appl Microbiol 93:463–472
Ferree MA, Shannon RD (2001) Evaluation of a second derivative UV/visible spectroscopy technique for nitrate and total nitrogen analysis of wastewater samples. Water Res 35:327–332
Fournier D, Halasz A, Spain J, Fiurasek P, Hawari J (2002) Determination of key metabolites during biodegradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine with Rhodococcus sp. strain DN22. Appl Environ Microbiol 68:166–172
Fuller ME, Perreault N, Hawari J (2010) Microaerophilic degradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) by three Rhodococcus strains. Lett Appl Microbiol 51:313–318
Hawari J, Beaudet S, Halasz A, Thiboutot S, Ampleman G (2000) Microbial degradation of explosives: biotransformation versus mineralization. Appl Microbiol Biotechnol 54:605–618
Indest KJ, Crocker FH, Athow R (2007) A TaqMan polymerase chain reaction method for monitoring RDX-degrading bacteria based on the xplA functional gene. J Microbiol Methods 68:267–274
Indest KJ, Jung CM, Chen HP, Hancock D, Florizone C, Eltis LD, Crocker FH (2010) Functional characterization of pGKT2, a 182-kilobase plasmid containing the xplAB genes, which are involved in the degradation of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine by Gordonia sp. strain KTR9. Appl Environ Microbiol 76:6329–6337
Jackson RJ, Rylott EL, Fournier D, Hawari J, Bruce NC (2007) Exploring the biochemical properties and remediation applications of the unusual explosive-degrading P450 system xplA/B. Proc Natl Acad Sci USA 104:16822–16827
Martínková L, Uhnáková B, Pátek M, Nesvera J, Kren V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177
Nejidat A, Kafka L, Tekoah Y, Ronen Z (2008) Effect of organic and inorganic nitrogenous compounds on RDX degradation and cytochrome P450 expression in Rhodococcus strain YH1. Biodegradation 19:313–320
Pennington JC, Brannon JM, Gunnison D, Harrelson DW, Zakikhani M, Miyares P, Jenkins TF, Clarke J, Hayes C, Ringleberg D, Perkins E, Fredrickson H (2001) Monitored natural attenuation of explosives. Soil Sediment Contam 10:45–70
Priestley JT, Coleman NV, Duxbury T (2006) Growth rate and nutrient limitation affect the transport of Rhodococcus sp. strain DN22 through sand. Biodegradation 17:571–576
Roh H, Yu C, Fuller M, Chu K-H (2009) Identification of hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine-degrading microorganisms via 15N-stable isotope probing. Environ Sci Technol 43:2505–2511
Ronen Z, Brenner A, Abeliovich A (1998) Biodegradation of RDX-contaminated wastes in a nitrogen-deficient environment. Water Sci Technol 38:219–224
Ronen Z, Yanovich Y, Goldin R, Adar E (2008) Metabolism of the explosive hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) in a contaminated vadose zone. Chemosphere 73:1492–1498
Rylott EL, Jackson RG, Sabbadin F, Seth-Smith HMB, Edwards J, Chong CS, Strand SE, Grogan G, Bruce NC (2011) The explosive-degrading cytochrome P450 XplA: Biochemistry, structural features and prospects for bioremediation. BBA Proteins Proteomics 1814:230–236
Seth-Smith HMB, Rosser SJ, Basran A, Travis ER, Dabbs ER, Nicklin S, Bruce NC (2002) Cloning, sequencing, and characterization of the hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl Environ Microbiol 68:4764–4771
Seth-Smith HMB, Edwards J, Rosser SJ, Rathbone DA, Bruce NC (2008) The explosive-degrading cytochrome P450 system is highly conserved among strains of Rhodococcus spp. Appl Environ Microbiol 74:4550–4552
Shen J, He R, Wang L, Zhang J, Zuo Y, Li Y, Sun X, Li J, Han W (2009) Biodegradation kinetics of picric acid by Rhodococcus sp. NJUST16 in batch reactors. J Hazard Mater 167:193–198
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tekoah Y, Abeliovich A, Nejidat A (1999) Participation of cytochrome P450 in the biodegradation of RDX by a Rhodococcus strain, p. 7. In: 2nd International Symposium, Biodegradation of Nitroaromatic Compounds and Explosives Leesburg, VA
US EPA (1997) Method 8330, SW-846 update III, part 4: 1 (B). US EPA, Office of Solid Waste, Washington, DC
US EPA (2006) Edition of the drinking water standards and health advisories. Office of Water, Washington, DC
Wani AH, Davis JL (2003) RDX biodegradation column study: influence of ubiquitous electron acceptors on anaerobic biotransformation of RDX. J Chem Technol Biotechnol 78:1082–1092
Acknowledgments
This work was supported in part by the Israel Water Authority and by the Eshkol Fellowship of Israel Ministry of Science and the Israel Science Foundation grant 167/08.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bernstein, A., Adar, E., Nejidat, A. et al. Isolation and characterization of RDX-degrading Rhodococcus species from a contaminated aquifer. Biodegradation 22, 997–1005 (2011). https://doi.org/10.1007/s10532-011-9458-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-011-9458-0