Abstract
Bioremediation is becoming an increasingly popular approach for the remediation of sites contaminated with the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX). Multiple lines of evidence are often needed to assess the success of such approaches, with molecular studies frequently providing important information on the abundance of key biodegrading species. Towards this goal, the current study utilized shotgun sequencing to determine the abundance and diversity of functional genes (xenA, xenB, xplA, diaA, pnrB, nfsI) and species previously associated with RDX biodegradation in groundwater before and after biostimulation at an RDX-contaminated Navy Site. For this, DNA was extracted from four and seven groundwater wells pre- and post-biostimulation, respectively. From a set of 65 previously identified RDX degraders, 31 were found within the groundwater samples, with the most abundant species being Variovorax sp. JS1663, Pseudomonas fluorescens, Pseudomonas putida, and Stenotrophomonas maltophilia. Further, 9 RDX-degrading species significantly (p<0.05) increased in abundance following biostimulation. Both the sequencing data and qPCR indicated that xenA and xenB exhibited the highest relative abundance among the six genes. Several genes (diaA, nsfI, xenA, and pnrB) exhibited higher relative abundance values in some wells following biostimulation. The study provides a comprehensive approach for assessing biomarkers during RDX bioremediation and provides evidence that biostimulation generated a positive impact on a set of key species and genes.
Key points
• A co-occurrence network indicated diverse RDX degraders.
• >30 RDX-degrading species were detected.
• Nine RDX-degrading species increased following biostimulation.
• Sequencing and high-throughput qPCR indicated that xenA and xenB were most abundant.






Similar content being viewed by others
Data availability
MG-RAST ID numbers and sequencing data have been summarized (Table S1), and the datasets are publicly available on MG-RAST.
References
Agency for Toxic Substances and Disease Registry (ATSDR) (2012) Toxicological profile for RDX.
Andeer PF, Stahl DA, Bruce NC, Strand SE (2009) Lateral transfer of genes for hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) degradation. Appl Environ Microbiol 75(10):3258–3262. https://doi.org/10.1128/AEM.02396-08
Arcadis (2004) In-situ enhanced anaerobic explosive treatment field test report. Milan Army Ammunition Plant Milan, Tennessee. Arcadis, Knoxville
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinform J 7:1–8. https://doi.org/10.2174/1875036201307010001
Bastian M, Heymann S, Jacomy M (2006) Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media
Bates D, Maechler M, Davis TA, Oehlschlägel J, Riedy J (2019) Matrix: sparse and dense matrix classes and methods.
Bernstein A, Adar E, Nejidat A, Ronen Z (2011) Isolation and characterization of RDX-degrading Rhodococcus species from a contaminated aquifer. Biodegradation 22(5):997–1005. https://doi.org/10.1007/s10532-011-9458-0
Bhushan B, Halasz A, Spain JC, Hawari J (2002) Diaphorase catalyzed biotransformation of RDX via N-denitration mechanism. Biochem Biophys Res Commun 296(4):779–784. https://doi.org/10.1016/s0006-291x(02)00874-4
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bryant C, DeLuca M (1991) Purification and characterization of an oxygen-insensitive NAD(P)H nitroreductase from Enterobacter cloacae. J Biol Chem 266(7):4119–4125. https://doi.org/10.1016/S0021-9258(20)64294-6
Bryant C, Hubbard L, McElroy WD (1991) Cloning, nucleotide sequence, and expression of the nitroreductase gene from Enterobacter cloacae. J Biol Chem 266(7):4126–4130. https://doi.org/10.1016/S0021-9258(20)64295-8
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59–60. https://doi.org/10.1038/nmeth.3176
Caballero A, Lazaro JJ, Ramos JL, Esteve-Nunez A (2005) PnrA, a new nitroreductase-family enzyme in the TNT-degrading strain Pseudomonas putida JLR11. Environ Microbiol 7(8):1211–1219. https://doi.org/10.1111/j.1462-2920.2005.00801.x
Chakraborty S, Sakka M, Kimura T, Sakka K (2008) Cloning and expression of a Clostridium kluyveri gene responsible for diaphorase activity. Biosci Biotechnol Biochem 72(3):735–741. https://doi.org/10.1271/bbb.70606
Coleman NV, Nelson DR, Duxbury T (1998) Aerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol Biochem 30(8-9):1159–1167
Collier JM, Chai B, Cole JR, Michalsen MM, Cupples AM (2019) High throughput quantification of the functional genes associated with RDX biodegradation using the SmartChip real-time PCR system. Appl Microbiol Biotechnol 103(17):7161–7175. https://doi.org/10.1007/s00253-019-10022-x
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11(485):1–6
Crocker FH, Indest KJ, Jung CM, Hancock DE, Fuller ME, Hatzinger PB, Vainberg S, Istok JD, Wilson E, Michalsen MM (2015) Evaluation of microbial transport during aerobic bioaugmentation of an RDX-contaminated aquifer. Biodegradation 26(6):443–451. https://doi.org/10.1007/s10532-015-9746-1
Csardi G, Nepusz T (2006) The igraph software package for complex network research. Int J Complex Syst 1695
EPA (2015) Third five-year review for Cornhusker Army Ammunition Plant Grand Island, Nebraska. Environmental Protection Agency Region 7, Lenexa
EPA (2017) Technical fact sheet – Hexahydro-1,3,5-trinitro1,3,5-triazine (RDX).
Felt D, Johnson JL, Larson S, Hubbard B, Henry K, Nestler C, Ballard JH (2013) Evaluation of treatment technologies for wastewater from insensitive munitions production. The US Army Engineer Research and Development Center
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. https://doi.org/10.3389/fmicb.2013.00291
Fuller ME, McClay K, Hawari J, Paquet L, Malone TE, Fox BG, Steffan RJ (2009) Transformation of RDX and other energetic compounds by xenobiotic reductases XenA and XenB. Appl Microbiol Biotechnol 84(3):535–544. https://doi.org/10.1007/s00253-009-2024-6
Fuller ME, McClay K, Higham M, Hatzinger PB, Steffan RJ (2010a) Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) bioremediation in groundwater: are known RDX-degrading bacteria the dominant players? Bioremediat J 14(3):121–134. https://doi.org/10.1080/10889868.2010.495367
Fuller ME, Perreault N, Hawari J (2010b) Microaerophilic degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by three Rhodococcus strains. Lett Appl Microbiol 51(3):313–318. https://doi.org/10.1111/j.1472-765X.2010.02897.x
Harrell FE (2020) Hmisc: harrell miscellaneous.
https://cumulis.epa.gov/supercpad/CurSites/srchsites.cfm. Accessed 2020
Huson DH, Beier S, Flade I, Gorska A, El-Hadidi M, Mitra S, Ruscheweyh HJ, Tappu R (2016) MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12(6):e1004957. https://doi.org/10.1371/journal.pcbi.1004957
Indest KJ, Crocker FH, Athow R (2007) A TaqMan polymerase chain reaction method for monitoring RDX-degrading bacteria based on the xplA functional gene. J Microbiol Methods 68(2):267–274. https://doi.org/10.1016/j.mimet.2006.08.008
Jugnia L-B, Manno D, Dodard S, Greer CW, Hendry M (2019) Manipulating redox conditions to enhance in situ bioremediation of RDX in groundwater at a contaminated site. Sci Total Environ 676:368–377. https://doi.org/10.1016/j.scitotenv.2019.04.045
Kahng H-Y, Lee B-U, Cho Y-S, Oh K-H (2007) Purification and characterization of the NAD(P)H-nitroreductase for the catabolism of 2,4,6-trinitrotoluene (TNT) in Pseudomonas sp. HK-6. Biotechnol Bioprocess Eng 12:433–440. https://doi.org/10.1007/BF02931067
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M (2016) KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44(D1):D457–D462. https://doi.org/10.1093/nar/gkv1070
Katoh K, Rozewicki J, Yamada KD (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 20(4):1160–1166. https://doi.org/10.1093/bib/bbx108
Kitts CL, Green CE, Otley RA, Alvarez MA, Unkefer PJ (2000) Type I nitroreductases in soil Enterobacteria reduce TNT (2,4,6,-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can J Microbiol 46(3):278–282. https://doi.org/10.1139/w99-134
Kwon MJ, O'Loughlin EJ, Antonopoulos DA, Finneran KT (2011) Geochemical and microbiological processes contributing to the transformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) in contaminated aquifer material. Chemosphere 84(9):1223–1230. https://doi.org/10.1016/j.chemosphere.2011.05.027
Lee BU, Choi MS, Kim DM, Oh KH (2017) Genome shuffling of Stenotrophomonas maltophilia OK-5 for improving the degradation of explosive RDX (Hexahydro-1,3,5-trinitro-1,3,5-triazine). Curr Microbiol 74(2):268–276. https://doi.org/10.1007/s00284-016-1179-5
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47(W1):W256–W259. https://doi.org/10.1093/nar/gkz239
Li RW, Giarrizzo JG, Wu S, Li W, Duringer JM, Craig AM (2014) Metagenomic insights into the RDX-degrading potential of the ovine rumen microbiome. PLoS One 9(11):e110505. https://doi.org/10.1371/journal.pone.0110505
Livermore JA, Jin YO, Arnseth RW, Lepuil M, Mattes TE (2013) Microbial community dynamics during acetate biostimulation of RDX-contaminated groundwater. Environ Sci Technol 47(14):7672–7678. https://doi.org/10.1021/es4012788
Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA (2008) The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. https://doi.org/10.1186/1471-2105-9-386
Michalsen MM, Weiss R, King A, Gent D, Medina VF, Istok JD (2013) Push-pull tests for estimating RDX and TNT degradation rates in groundwater. Ground Water Monit Remediat 33(3):61–68. https://doi.org/10.1111/gwmr.12016
Michalsen MM, King AS, Rule RA, Fuller ME, Hatzinger PB, Condee CW, Crocker FH, Indest KJ, Jung CM, Istok JD (2016) Evaluation of biostimulation and bioaugmentation to stimulate hexahydro-1,3,5-trinitro-1,3,5,-triazine degradation in an aerobic groundwater aquifer. Environ Sci Technol 50(14):7625–7632. https://doi.org/10.1021/acs.est.6b00630
Michalsen MM, King AS, Istok JD, Crocker FH, Fuller ME, Kucharzyk KH, Gander MJ (2020) Spatially-distinct redox conditions and degradation rates following field-scale bioaugmentation for RDX-contaminated groundwater remediation. J Hazard Mater 387:121529. https://doi.org/10.1016/j.jhazmat.2019.121529
Nejidat A, Kafka L, Tekoah Y, Ronen Z (2008) Effect of organic and inorganic nitrogenous compounds on RDX degradation and cytochrome P-450 expression in Rhodococcus strain YH1. Biodegradation 19(3):313–320
Papadopoulos JS, Agarwala R (2007) COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23(9):1073–1079. https://doi.org/10.1093/bioinformatics/btm076
Parks DH, Tyson GW, Hugenholtz P, Beiko RG (2014) STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30(21):3123–3124. https://doi.org/10.1093/bioinformatics/btu494
Peterson FJ, Mason RP, Hovsepian J, Holtzman JL (1979) Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J Biol Chem 254(10):4009–4014
Pruitt KD, Tatusova T, Maglott DR (2005) NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 33(Database issue):D501–D504. https://doi.org/10.1093/nar/gki025
R Core Team (2018) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Rho M, Tang H, Ye Y (2010) FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38(20):e191. https://doi.org/10.1093/nar/gkq747
Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Loffler FE (2006) Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 72(4):2765–2774. https://doi.org/10.1128/AEM.72.4.2765-2774.2006
RStudio Team (2020) RStudio: Integrated Development for R. RStudio, PBC, Boston
Rylott EL, Jackson RG, Sabbadin F, Seth-Smith HMB, Edwards J, Chong CS, Strand SE, Grogan G, Bruce NC (2011) The explosive-degrading cytochrome P450 XplA: biochemistry, structural features and prospects for bioremediation. Bba-Pro Proteom 1814(1):230–236. https://doi.org/10.1016/j.bbapap.2010.07.004
Seth-Smith HM, Rosser SJ, Basran A, Travis ER, Dabbs ER, Nicklin S, Bruce NC (2002) Cloning, sequencing, and characterization of the hexahydro-1,3,5-Trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl Environ Microbiol 68(10):4764–4771. https://doi.org/10.1128/aem.68.10.4764-4771.2002
Seth-Smith HMB, Edwards J, Rosser SJ, Rathbone DA, Bruce NC (2008) The explosive-degrading cytochrome P450 system is highly conserved among strains of Rhodococcus spp. Appl Environ Microbiol 74(14):4550–4552
Sherrill-Mix S (2019) taxonomizr: functions to work with NCBI accessions and taxonomy
Stedtfeld RD, Williams MR, Fakher U, Johnson TA, Stedtfeld TM, Wang F, Khalife WT, Hughes M, Etchebarne BE, Tiedje JM, Hashsham SA (2016) Antimicrobial resistance dashboard application for mapping environmental occurrence and resistant pathogens. FEMS Microbiol Ecol 92(3). https://doi.org/10.1093/femsec/fiw020
Thompson KT, Crocker FH, Fredrickson HL (2005) Mineralization of the cyclic nitramine explosive hexahydro-1,3,5-trinitro-1,3,5-triazine by Gordonia and Williamsia spp. Appl Environ Microbiol 71(12):8265–8272. https://doi.org/10.1128/Aem.71.12.8265-8272.2005
Wang D, Boukhalfa H, Marina O, Ware DS, Goering TJ, Sun F, Daligault HE, Lo CC, Vuyisich M, Starkenburg SR (2017) Biostimulation and microbial community profiling reveal insights on RDX transformation in groundwater. Microbiologyopen 6(2). https://doi.org/10.1002/mbo3.423
Wani AH, O'Neal BR, Davis JL, Hansen LD (2002) Treatability study for biologically active zone enhancement (BAZE) for in situ RDX degradation in groundwater. U.S. Army Engineer Research and Development Center, Vicksburg
Wickham H (2016) ggplot2: elegant graphics for data analysis.
Wilson FP, Cupples AM (2016) Microbial community characterization and functional gene quantification in RDX-degrading microcosms derived from sediment and groundwater at two naval sites. Appl Microbiol Biotechnol 100(16):7297–7309. https://doi.org/10.1007/s00253-016-7559-8
Acknowledgements
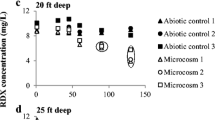
Thanks to M. M. Michalsen (U.S. Army Engineer Research Development Center) for providing the groundwater and the RDX concentrations.
Funding
Thanks to Malcolm Gander (Naval Facilities Engineering Command) for partially funding this project and thanks to Craig Tobias (University of Connecticut) for facilitating project funding.
Author information
Authors and Affiliations
Contributions
Hongyu Dang: Conceptualization, methodology, programming, validation, formal analysis, writing, and investigation. Alison Cupples: Resources, data curation, writing-review and editing, visualization, supervision, project administration, and funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical statement
This work was, in part, supported by Naval Facilities Engineering Command.
Human and animal rights and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 4679 kb)
Rights and permissions
About this article
Cite this article
Dang, H., Cupples, A.M. Diversity and abundance of the functional genes and bacteria associated with RDX degradation at a contaminated site pre- and post-biostimulation. Appl Microbiol Biotechnol 105, 6463–6475 (2021). https://doi.org/10.1007/s00253-021-11457-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11457-x