Abstract
Aquatic invasive plant species cause negative impacts to economies and ecosystems worldwide. Traditional survey methods, while necessary, often do not result in timely detections of aquatic invaders, which can be cryptic, difficult to identify, and exhibit very rapid growth and reproduction rates. Environmental DNA (eDNA) is a relatively new method that has been used to detect multiple types of animals in freshwater and marine ecosystems through tissues naturally shed from the organism into the water column or sediment. While eDNA detection has proven highly effective in the detection of aquatic animals, we know less about the efficacy of eDNA as an effective surveillance tool for aquatic plants. To address this disparity, we designed mesocosm experiments with Elodea species to determine the ability to detect accumulation and degradation of the DNA signal for aquatic plants, followed by field surveillance of the highly invasive Hydrilla verticillata in freshwaters across several U.S. geographic regions. In both lab and field experiments, we designed a high sensitivity quantitative PCR assay to detect the aquatic plant species. In both experiments, plant eDNA detection was successful; we saw accumulation of DNA when plants were introduced to tanks and a decrease in DNA over time after plants were removed. We detected eDNA in the field in areas of known Hydrilla distribution. Employing eDNA detection for aquatic plants will strengthen efforts for early detection and rapid response of invaders in global freshwater ecosystems.



Similar content being viewed by others
References
Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM (2014) Environmental conditions influence eDNA persistence in aquatic systems. Environ Sci Technol 48(3):1819–1827
Beja-Pereira A, Oliveira R, Alves PC, Schwartz MK, Luikart G (2009) Advancing ecological understandings through technological transformations in noninvasive genetics. Mol Ecol Resour 9(5):1279–1301
Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Douglas WY, de Bruyn M (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29(6):358–367
Cook CD, Lüönd R (1982) A revision of the genus Hydrilla (Hydrocharitaceae). Aquat Bot 13:485–504
Cowart DA, Renshaw MA, Gantz CA, Umek J, Chandra S, Egan SP, Lodge DM, Larson ER (2018) Development and field validation of an environmental DNA (eDNA) assay for the invasive Asian clam, Corbicula fluminea (Müller, 1774). Manag Biol Invasion 9(1):27–37
Darling JA, Mahon AR (2011) From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ Res 111(7):978–988
Deiner K, Altermatt F (2014) Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 9(2):e88786
Dougherty MM, Larson ER, Renshaw MA, Gantz CA, Egan SP, Erickson DM, Lodge DM (2016) Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. J Appl Ecol 53(3):722–732
Egan SP, Barnes MA, Hwang CT, Mahon AR, Feder JL, Ruggiero ST, Tanner CE, Lodge DM (2013) Rapid invasive species detection by combining environmental DNA with light transmission spectroscopy. Conserv Lett 6(6):402–409
Egan SP, Grey E, Olds B, Feder JL, Ruggiero ST, Tanner CE, Lodge DM (2015) Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environ Sci Technol 49(7):4113–4121
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425
Fisher E (2015) GLP update: Indiana Hydrilla eradication & Starry stonewort battle. Update to the Great Lakes Panel. http://glc.org/files/projects/ais/GLPMeeting-April2015-Fisher-SSW.pdf. Accessed 19 July 2016
Foote AD, Thomsen PF, Sveegaard S, Wahlberg M, Kielgast J, Kyhn LA, Salling AB, Galatius A, Orlando L, Gilbert MTP (2012) Investigating the potential use of environmental DNA (eDNA) for genetic monitoring of marine mammals. PLoS ONE 7(8):e41781
Fujiwara A, Matsuhashi S, Doi H, Yamamoto S, Minamoto T (2016) Use of environmental DNA to survey the distribution of an invasive submerged plant in ponds. Freshw Sci 35(2):748–754
Global Invasive Species Database (2016) Species profile: Hydrilla verticillata. http://www.iucngisd.org/gisd/species.php?sc=272. Accessed 21 July 2016
Gu W, Swihart RK (2004) Absent or undetected? Effects of non-detection of species occurrence on wildlife–habitat models. Biol Conserv 116(2):195–203
Gunawardana M, Chang S, Jimenez A, Holland-Moritz D, Holland-Moritz H, La Val TP, Lund C, Mullen M, Olsen J, Sztain TA, Yoo J (2014) Isolation of PCR quality microbial community DNA from heavily contaminated environments. J Microbiol Methods 102:1–7
Hollingsworth PM, Graham SW, Little DP (2011) Choosing and using a plant DNA barcode. PLoS ONE 6(5):e19254
Hulme PE (2012) Weed risk assessment: a way forward or a waste of time? J Appl Ecol 49(1):10–19
Jacono CC, Richerson MM, Morgan VH, Pfingsten IA (2015) Hydrilla verticillata. USGS Nonindigenous Aquatic Species Database, Gainesville, FL. http://nas.er.usgs.gov/queries/FactSheet.aspx?SpeciesID=6. Accessed 21 July 2016
Jane SF, Wilcox TM, McKelvey KS, Young MK, Schwartz MK, Lowe WH, Letcher BH, Whiteley AR (2015) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15(1):216–227
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv Lett 4:150–157
Jerde CL, Chadderton WL, Mahon AR, Renshaw MA, Corush J, Budny ML, Mysorekar S, Lodge DM (2013) Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can J Fish Aquat Sci 70(4):522–526
Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L (1998) COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377(1–3):147–159
Kratville D (2013) The California Department of Food and Agriculture Hydrilla eradication program annual progress report 2013. California Department of Food and Agriculture, Sacramento, CA. https://www.cdfa.ca.gov/plant/ipc/hydrilla/pdfs/2013HydrillaAnnualReport.pdf. Accessed 15 September 2015
Lake County Department of Public Works, Water Resources Division (2004) Clear lake integrated aquatic plant management plan. http://www.co.lake.ca.us/Assets/WaterResources/Aquatic+Plant+Management+Areas/Aquatic+Plant+Management+Plan.pdf. Accessed 15 Sept 2015
Langeland KA (1996) Hydrilla verticillata (LF) Royle (Hydrocharitaceae), “The Perfect Aquatic Weed”. Castanea 61(3):293–304
Larson ER, Renshaw MA, Gantz CA, Umek J, Chandra S, Lodge DM, Egan SP (2017) Using environmental DNA (eDNA) to survey for the crayfishes Orconectes rusticus and Pacifastacus leniusculus between their reciprocal invasive ranges in North America. Hydrobiologia 800:173–185
Lodge DM, Williams S, MacIsaac HJ, Hayes KR, Leung B, Reichard S, Mack RN, Moyle PB, Smith M, Andow DA, Carlton JT (2006) Biological invasions: recommendations for US policy and management. Ecol Appl 16(6):2035–2054
Lodge DM, Turner CR, Jerde CL, Barnes MA, Chadderton L, Egan SP, Feder JL, Mahon AR, Pfrender ME (2012) Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Mol Ecol 21(11):2555–2558
Lodge DM, Simonin PW, Burgiel SW, Keller RP, Bossenbroek JM, Jerde CL, Kramer AM, Rutherford ES, Barnes MA, Wittmann ME, Chadderton WL, Apriesnig JL, Beletsky D, Cooke R, Drake JM, Egan SP, Finnoff DC, Gantz CA, Grey EK, Hoff MH, Howeth JG, Jensen RA, Larson ER, Mandrak NE, Mason DM, Martinez FA, Newcomb TJ, Rothlisberger JD, Tucker AJ, Warziniack TW, Zhang H (2016) Risk analysis and bioeconomics of invasive species to inform policy and management. Annu Rev Environ Resour 41:453–488
Longmire JL, Maltbie M, Baker RJ (1997) Use of “lysis buffer” in DNA isolation and its implication for museum collections. Mus Tex Tech Univ 163:1–3
Lovell SJ, Stone SF, Fernandez L (2006) The economic impacts of aquatic invasive species: a review of the literature. Agric Resour Econ Rev 35:195–208
Matsuhashi S, Doi H, Fujiwara A, Watanabe S, Minamoto T (2016) Evaluation of the environmental DNA method for estimating distribution and biomass of submerged aquatic plants. PLoS ONE 11(6):e0156217
Minamoto T, Uchii K, Takahara T, Kitayoshi T, Tsuji S, Yamanaka H (2017) Nuclear internal transcribed spacer-1 as a sensitive genetic marker for environmental DNA studies in common carp Cyprinus carpio. Mol Ecol Resour 17(2):324–333
Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D (2007) Release and persistence of extracellular DNA in the environment. Environ Biosaf Res 6:37–53
Office of Technology Assessment (OTA) (1993) Harmful non-indigenous species in the United States. Publication No. OTA-F-565, OTA, U.S. Congress, Washington, DC
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2013) Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can J Fish Aquat Sci 70(8):1123–1130
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2014) Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol Ecol Resour 14:109–116
Pimentel D, Zuniga R, Morrison D (2005) Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ 52(3):273–288
Rees HC, Maddison BC, Middleditch DJ, Patmore JR, Gough KC (2014) Review: the detection of aquatic animal species using environmental DNA—a review of eDNA as a survey tool in ecology. J Appl Ecol 51(5):1450–1459
Renshaw MA, Olds BP, Jerde CL, McVeigh MM, Lodge DM (2015) The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol–chloroform–isoamyl alcohol DNA extraction. Mol Ecol Resour 15(1):168–176
Rogers SO, Bendich AJ (1987) Ribosomal RNA genes in plants: variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
SAS Institute Inc (2013) Using JMP 11. SAS Institute Inc, Cary
Schmelzle MC, Kinziger AP (2016) Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol Ecol Resour 16:895–908
Schmidt BR, Kéry M, Ursenbacher S, Hyman OJ, Collins JP (2013) Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods Ecol Evol 4:646–653
Scriver M, Marinich A, Wilson C, Freeland J (2015) Development of species-specific environmental DNA (eDNA) markers for invasive aquatic plants. Aquat Bot 122:27–31
Shadel GS, Clayton DA (1997) Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem 66(1):409–435
Smith CJ, Osborn AM (2009) Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol Ecol 67:6–20
Spear SF, Groves JD, Williams LA, Waits LP (2015) Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol Conserv 183:38–45
Strayer DL (2010) Alien species in fresh waters: ecological effects, interactions with other stressors, and prospects for the future. Freshw Biol 55(s1):152–174
Strayer DL, Eviner VT, Jeschke JM, Pace ML (2006) Understanding the long-term effects of species invasions. Trends Ecol Evol 21(11):645–651
Strickler KM, Fremier AK, Goldberg CS (2015) Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol Conserv 183:85–92
Svec D, Tichopad A, Novosadova V, Pfaffl MW, Kubista M (2015) How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol Detect Quantif 3:9–16
Thomsen PF, Willerslev E (2015) Environmental DNA—an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18
Trebitz A, Hoffman J, Darling J, Pilgrim E, Kelly J, Schardt J, Brown E, Chadderton L, Egan SP, Grey E, Hashsham S, Klymus K, Mahon A, Ram J, Schultz M, Stepien C (2017) Science status and needs for implementing early detection monitoring of aquatic non-indigenous species. J Environ Manag 202:299–310
UNEP (2006) Africa Environment Outlook 2. Division of Early Warning and Assessment, United Nations Environment Programme, Nairobi
Valentini A, Pompanon F, Taberlet P (2009) DNA barcoding for ecologists. Trends Ecol Evol 24(2):110–117
Vander Zanden MJ, Hansen GJ, Higgins SN, Kornis MS (2010) A pound of prevention, plus a pound of cure: early detection and eradication of invasive species in the Laurentian Great Lakes. J Great Lakes Res 36(1):199–205
Yoccoz NG (2012) The future of environmental DNA in ecology. Mol Ecol 21:2031–2038
Acknowledgements
We would like to thank Haley Erickson and Eric Larson for field assistance and Lizz Radican and Bill Wang for laboratory assistance with this project. Kelley Morris, Cullen Ondracek, Mark Webb from Texas Parks and Wildlife helped with Hydrilla field sites in Texas. Kristen Heyer and Bill Hamilton of the Maryland DNR provided the boat and sampling assistance at Mattawoman Creek. Joseph Love, Bruce Michael, and John Mullican of the Maryland DNR provided sampling coordinates and advice for sampling. Ryan Argo, Ohio River Valley Water Sanitation Commission (ORSANCO), provided location coordinates for sampling in the Ohio River. Eric Fischer, Indiana Department of Natural Resources, assisted with collection permits and information about Lake Manitou. Sudeep Chandra provided information about Clear Lake in California. John Madsen (USDA) provided Hydrilla tissue for the assay. Linyi Zhang created the map figures. Partial support was provided to CAG by the Strecker Lab at Portland State University. This research was supported by US Environmental Protection Agency Grant EPA-R5-GL2012-1 to SPE and DML and Biotechnology Risk Assessment Grant Program Competitive Grant Nos. 2013-33522-21007 and 2016-33522-25629 to SPE from the USDA National Institute of Food and Agriculture and the Agricultural Research Service. Angela Strecker, Meredith Holgerson, and Ariana Chiapella provided helpful comments on earlier drafts of the manuscript. We would especially like to thank the associate editor and two anonymous reviewers for their thoughtful attention to this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material (Online Resource 1)
Details of primer design for Elodea canadensis, Elodea nuttallii, and Hydrilla verticillata. Details of extraction protocol for eDNA samples. Copy number details for matK data for mesocosm experiment. (DOCX 1348 kb)
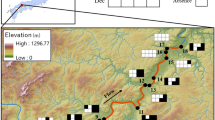
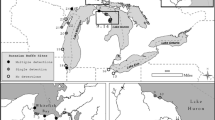
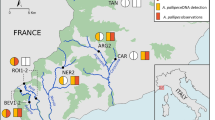
Appendix 1: Sampling locations for all field sites in this study
Appendix 1: Sampling locations for all field sites in this study
See Table 3.
Rights and permissions
About this article
Cite this article
Gantz, C.A., Renshaw, M.A., Erickson, D. et al. Environmental DNA detection of aquatic invasive plants in lab mesocosm and natural field conditions. Biol Invasions 20, 2535–2552 (2018). https://doi.org/10.1007/s10530-018-1718-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-018-1718-z