Abstract
Monitoring, management and conservation of rare and elusive species often requires early detection of individuals, especially for re-introduced and endangered taxa. Environmental DNA (eDNA) approaches can enhance the detection power of traditional biomonitoring methods for low-density, newly-established populations. In this study, we used species-specific Real Time PCR TaqMan assays to assess the presence of two endangered freshwater species, the white-clawed crayfish Austropotamobius pallipes and the Eurasian otter Lutra lutra at eight sites in four river catchments in Liguria (northwestern Italy). The Eurasian otter was considered extinct in the study area since the 1980s. However, recent, although scattered sightings indicated a recolonisation by a few individuals. The white-clawed crayfish populations declined drastically and became increasingly dispersed in the western part of Liguria. Our eDNA analysis confirmed the presence of both species in some of the selected rivers and detected Eurasian otter DNA where the species was not recorded through traditional monitoring methods. This study confirms eDNA-based monitoring approaches as valuable tools to assess the presence of rare and elusive species and help implement protection plans at a local scale.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodiversity monitoring is an essential tool to understand species distribution and inform wildlife conservation strategies. The effective management of rare and elusive species often requires early detection of populations, especially for reintroduced and endangered taxa (Maxwell and Jennings 2005; Jerde et al. 2011; Deiner et al. 2021; Peralta et al. 2023). The analysis of environmental DNA (eDNA) can complement traditional biomonitoring methods resulting in a more detailed description of community composition, especially in aquatic environments (Tsuji et al. 2019).
Monitoring methods based on eDNA allow for early detection of populations in which individuals are either at low densities, early ontogenetic or cryptic stages. eDNA-based techniques can help identify different species from a single sample (metabarcoding eDNA) or record the presence of a species of interest (targeted eDNA). In particular, probe-based Real Time PCR is now widely used to detect presence of a target species in an eDNA sample (Taberlet et al. 2018; Xia et al. 2018; Pawlowski et al. 2020).
Inland waters and freshwater ecosystems are among the most threatened environments and are experiencing high rates of biodiversity decline (Dudgeon 2019). The International Union for Conservation of Nature (IUCN) Red List reports that almost 22% of the known freshwater species are threatened (IUCN 2023; William-Subiza and Epele 2021). Two iconic endangered Italian native freshwater species are the Eurasian otter (Lutra lutra Linnaeus, 1758) and the white-clawed crayfish (Austropotamobius pallipes species complex Lereboullet, 1858).
Despite being found in a wide variety of aquatic environments in Asia, Europe and North Africa, the Eurasian otter is listed as Near-Threatened in the IUCN Red List. This species is listed as Endangered in Italy (Loy et al. 2020), where populations have been steadily declining since the last century due to habitat reduction and degradation (Loy 2018). Thanks to a number of conservation actions, otter populations are now recovering in some European countries, including southern and central Italy (Elmeros et al. 2006; Prigioni et al. 2007; Loy 2018; Bolinesi et al. 2019; Buglione et al. 2020a, b; Gaudiano et al. 2023). Recently, the presence of L. lutra has also been reported in northern Italian regions (Friuli Venezia Giulia, Veneto, Trentino Alto Adige and Liguria), probably as a result of recolonisation from neighbouring countries (Righetti 2011; Pavanello et al. 2015; Malthieux 2020; Nadai et al. 2022).
The white-clawed crayfish is threatened by habitat modification, pollution, competition with invasive alien species and the onset of lethal diseases (Holdich 2003). Many countries, including Italy, have experienced a massive population decline over the last few decades, so that this species has been listed as Endangered in the IUCN Red List since 2010 (Füreder et al. 2010). Italian populations of the white-clawed crayfish belong to two distinct species: A. pallipes, which occurs in the western part of Liguria, and A. italicus (including four subspecies, see Fratini et al. 2005), which is present in the rest of the Italian peninsula.
In this study, we used a Real Time PCR Taqman assay to assess the presence of the white-clawed crayfish and the Eurasian otter in four river catchments in Liguria (northwestern Italy). The Eurasian otter has been considered extinct in this region since the 1980s (see Prigioni et al. 2007). However, an isolated population was recently recorded in adjacent areas (Malthieux 2020). The white-clawed crayfish populations have declined drastically in numbers and have become increasingly dispersed in western Liguria. In this context, an eDNA-based monitoring approach is an ideal, non-invasive tool to detect the presence of both species which presumably occur in the area at low-densities. Our study aims at defining the current distribution range of L. lutra and A. pallipes in western Liguria and informing future management and conservation plans.
Materials and methods
Study area and sampling
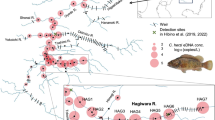
The study area included four river catchments in western Liguria (Italy): Roia-Bevera, Argentina, Nervia and Tanaro (Fig. 1). The Tanaro basin is located at the border of the Padano-Venetian ichthyogeographic district, while the other three catchments belong to the Tuscano-Latium ichthyogeographic zone. The Roia-Bevera basin is partially in French territory (Bianco 1990, 1995). Sampling was conducted in October 2022 in eight sites, two along the Roia river (ROI1 and ROI2), two in the Bevera river (BEV1 and BEV2), and one along the Argentina (ARG2), Tanaro (TAN), Carpasina (CAR) and Nervia (NER2) rivers (Fig. 1). Rivers and sampling sites were selected based on occurrence of L. lutra and A. pallipes recorded during previous monitoring campaigns (see Salvidio et al. 2002; Bologna and Cristiani 2012; Capurro et al. 2015; Malthieux 2020; Ottonello pers. obs.). Moreover, the study area represents the most direct route for recolonisation by otter populations from France (Malthieux 2020).
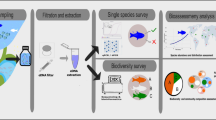
Water samples were collected at each site in six spatial replicates randomly selected on the two banks and in the middle of the river along a 150 m long transect. Each replicate consisted of one litre of water collected using plastic jars previously sterilised with sodium hypochlorite. Water samples were filtered on-site using a portable hand vacuum-pump connected to a polypropylene flask (Thermo Fisher Scientific). We used sterile disposable filter units with nitrocellulose membrane and a pore size of 0.2 μm (Thermo Fisher Scientific). All filters were immediately preserved in absolute ethanol upon water collection and stored at -20 °C prior to DNA extraction. A jar containing one litre of DNA-free deionized sterile water was left opened at each site for two minutes. The water was then filtered on-site and used as negative field control.
DNA extraction
Environmental DNA purification was carried out in a laminar flow cabinet using sterile equipment to avoid exogeneous DNA contamination. eDNA was extracted from nitrocellulose membrane filters using the ZymoBIOMICS DNA Miniprep Kit (Zymo Research) according to the manufacturer’s guidelines, eluted in a final volume of 50 µL of sterilised water and preserved at -20 °C.
To verify that DNA extraction from water samples provided sufficient metazoan genetic material, we measured DNA concentrations using the Qubit dsDNA HS Assay Kit (Invitrogen) and amplified two universal mitochondrial DNA (mtDNA) metabarcode markers using PCR primers Tele02 and Vert01 (Riaz et al. 2011; Taberlet et al. 2018). Amplification reactions were performed using 1X Invitrogen Taq DNA Polymerase PCR Buffer, 1 U of Taq DNA Polymerase (Thermo Fisher Scientific), 2 mM MgCl2, 0.3 mM dNTPs, 0.5 µM of each primer and 1 µL of sample DNA in 25 µL total volume. The following amplification conditions were used: 5 min at 94 °C, 35 cycles of 94 °C for 30 s, 54 °C (Tele02) or 49 °C (Vert01) for 30 s, 72 °C for 1 min and a final extension step at 72 °C for 10 min. PCR products were resolved on a 1.2% agarose gel stained with GelRed Nucleic Acid Gel Stain (Biotum). DNA-free ddH2O and L. lutra DNA extracted from fresh biological material were included in each PCR reaction as negative and positive controls, respectively.
Real time PCR assay
Presence of target species in the eDNA samples was detected using species-specific probe-based TaqMan assays. For L. lutra, we used the assay reported by Thomsen et al. (2012) which amplifies a 80 bp long fragment of the mtDNA cytochrome b gene. For A. pallipes, we used the assay reported by Troth et al. (2020) which amplifies a 109 bp fragment of the mtDNA cytochrome c oxidase subunit I gene. Each assay included two PCR primers and a target-specific dual labelled probe with fluorescent reporter and non-fluorescent quencher. A ROX fluorescent dye included in the Master Mix was used as an internal passive reference to normalize PCR fluorescent dye signals.
Positive controls consisted of L. lutra and A. pallipes DNA extracted from ethanol-preserved samples from the Natural History Museum of the University of Florence, Italy. For the white-clawed crayfish, we included positive controls of A. pallipes and three subspecies of A. italicus (A. i. carinthiacus, A. i. italicus and A. i. meridionalis). DNA extractions were performed from muscle tissues using the PureLink Genomic DNA Mini Kit (Thermo Fisher Scientific) following the manufacturer’s protocol.
Real Time PCR assays were performed on a QuantStudio 7 Flex Real-Time PCR System (Life Technologies). Ten-fold serial dilutions of positive control DNA were tested in ten replicates to assess Real Time PCR amplification efficiency and define the limit of detection (LOD, expressed as threshold cycle Ct) as reported in Bustin et al. (2009); Klymus et al. (2020a, b). Amplification conditions were as follows: a Pre-Read Stage at 60 °C for 30 s, hold at 95 °C for 20 s, 55 cycles at 95 °C for 1 s and 52 °C for 20 s followed by a Post-Read Stage at 60 °C for 30 s. Amplification reactions were conducted in a total volume of 20 µL containing 1X TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), 0.5 µM of each primer, 0.25 µM TaqMan probe and 5 µL template DNA.
For each sampling site, eDNA samples, positive and negative controls were amplified in technical triplicate in the same PCR run. Negative field controls were processed as eDNA samples. A Real Time PCR replicate was recorded as positive if Ct was lower than the LOD, it had a uniform curve morphology and no amplification occurred in the negative template and field controls (Bustin et al. 2009; Ficetola et al. 2015; Klymus et al. 2020a, b). Following the criteria proposed by Taberlet et al. (1996), Ficetola et al. (2015), Buxton et al. (2021) and Sanz et al. (2023) to avoid false positive and negative results, the presence of a species was considered as ascertained in the case of two positive amplifications out of the total of technical and spatial replicates.
Results and discussion
Environmental DNA concentrations ranged from 0.075 to 36.6 ng µL− 1. The Tele02 and Vert01 PCR assays confirmed successful extraction of metazoan DNA from the eDNA samples for we obtained a visible band for each sample when loaded on an agarose gel. Negative PCR controls excluded the possibility of false-positive amplification due to human DNA contamination.
For both detection assays, the amplification efficiency of Real Time PCR estimated by means of calibration curves determined using serial dilutions of the positive controls was about 90% (L. lutra: y = -3.63x + 24.05, r2 = 0.99; A. pallipes: y = -3.5x + 25.68, r2 = 0.97). The LOD corresponded to a Ct value of 43.7 ± 0.1 for the Eurasian otter assay and a Ct value of 43.3 ± 0.3 for the white-clawed crayfish assay. Ct values of positive Real Time PCR amplifications ranged from 37.7 to 43.5 for the otter assay and from 36.7 to 40.2 for the crayfish assay (Table 1).
Negative field and PCR controls produced no amplification, while PCR products of positive controls confirmed the reliability of the selected assays. For the white-clawed crayfish, we obtained a positive amplification for A. pallipes and the three tested subspecies of A. italicus. This result indicated that the sensitivity of the assay did not allow for the distinction between species and subspecies belonging to the A. pallipes species complex.
We recorded the presence of the Eurasian otter in four out of the six investigated rivers (Roia, Bevera, Argentina and Nervia). No presence of L. lutra was recorded in the Tanaro and Carpasina rivers. We detected the presence of the white-clawed crayfish in all river streams but the Tanaro and Roia rivers (Fig. 1; Table 1).
Historical sightings of the Eurasian otter in the western part of Liguria suggested the presence of few individuals in the high valley of the Tanaro, Argentina, Nervia, Roia and Bevera rivers (Balletto 1977; Vigna Taglianti and Bologna 1982). The species was considered extinct in the area ever since (Prigioni et al. 2007; Bologna and Cristiani 2012). Traces of L. lutra have been recently recorded upstream of BEV1-2 and ROI1-2 sampling sites in the Roia and Bevera rivers, indicating the presence of a relict population (Malthieux 2020). Our eDNA analysis confirmed the presence of the Eurasian otter on the Italian side of these rivers and detected the presence of L. lutra in adjacent basins where it was not previously observed (i.e., the Argentina and Nervia rivers).
The presence of the white-clawed crayfish was reported in five out of the six rivers included in this study, that is the Bevera, Nervia, Argentina, Carpasina (Capurro et al. 2015) and Tanaro (Salvidio et al. 2002). Recent observations, however, confirmed the presence of A. pallipes in all the above rivers but the Tanaro. Our eDNA approach confirmed recent surveys as we recorded the occurrence of the white-clawed crayfish only in BEV1, NER2, CAR and ARG2.
The majority of sites where we detected the presence of our target species are in close proximity to (NER2, ARG2, CAR) or mark the boundaries (BEV1-2) of Natura 2000 sites (Fig. 1). None of these Natura 2000 sites report L. lutra in their Standard Data Forms (SDF), while A. pallipes is only listed in the SDF of the protected areas established in the upper parts of the Carpasina and Argentina rivers.
This study confirms that eDNA-based monitoring approaches enhance the detection power of traditional biomonitoring surveys and are valuable tools to inform efficient management and protection schemes for threatened species (Pascher et al. 2022). Further investigations are needed to define in detail the distribution range of the target species in western Italy and reconstruct the recolonization routes of L. lutra of the Italian watersheds. Moreover, additional studies are needed to understand whether the presence of the Eurasian otter in western Italy is occasional or the species is reestablishing well-structured and viable populations.
Data availability
The data generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Balletto E (1977) Analisi faunistico-venatoria ed ecologica della Regione Liguria. Tipografia Don Bosco, Genova
Bianco PG (1990) Potential role of the palaeohistory of the Mediterranean and paratethys basins on the early dispersal of Euro-Mediterranean freshwater fishes. Ichthyol Explor Freshw 1:167–184
Bianco PG (1995) Mediterranean endemic freshwater fishes of Italy. Biol Conserv 72(2):159–170. https://doi.org/10.1016/0006-3207(94)00078-5
Bolinesi F, Viglietti S, Maio N, Guarino FM (2019) Presence of the eurasian otter Lutra lutra (Linnaeus, 1758) (Mammalia Mustelidae) in the Foce Sele-Tanagro Nature Reserve (Campania, Southern Italy). Biodivers J 10(2):121–126. https://doi.org/10.31396/Biodiv.Jour.2019.10.2.121.126
Bologna MA, Cristiani G (2012) Contributo alla teriofauna dell’atra Val Tanaro, Alpi Liguri (CN-IM). Riv Piemont Stor Nat 33:295–319
Buglione M, Petrelli S, Troiano C, Notomista T, Petrella A, De Riso L, Poerio L, Cascini V, Bartolomei R, Fulgione D (2020a) Spatial genetic structure in the eurasian otter (Lutra lutra) meta-population from its core range in Italy. Contrib Zool 90(1):70–92. https://doi.org/10.1163/18759866-BJA10012
Buglione M, Petrelli S, Troiano C, Notomista T, Rivieccio E, Fulgione D (2020b) The diet of otters (Lutra lutra) on the Agri river system, one of the most important presence sites in Italy: a molecular approach. PeerJ 8:e9606. https://doi.org/10.7717/peerj.9606
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. https://doi.org/10.1373/clinchem.2008.112797
Buxton A, Matechou E, Griffin J, Diana A, Griffiths RA (2021) Optimising sampling and analysis protocols in environmental DNA studies. Sci Rep 11(1):11637. https://doi.org/10.1038/s41598-021-91166-7
Capurro M, Oneto F, Ottonello D, Pericci R, Mignone W, Riina MV, Acuti PL (2015) Nota sulla presenza di esemplari xantocroici di gambero di fiume Austropotamobius pallipes specie complex (Lerebouillet, 1858) in Liguria (Italia nord-occidentale). Ann Mus Civ Stor Nat G Doria VIII:1–12
Deiner K, Yamanaka H, Bernatchez L (2021) The future of biodiversity monitoring and conservation utilizing environmental DNA. Environ DNA 3:3–7. https://doi.org/10.1002/edn3.178
Dudgeon D (2019) Multiple threats imperil freshwater biodiversity in the Anthropocene. Curr Biol 29(19):R960–R967. https://doi.org/10.1016/j.cub.2019.08.002
Elmeros M, Hammershoj M, Madsen A, Sogaard B (2006) Recovery of the otter Lutra lutra in Denmark monitored by field surveys and collection of carcasses. Hystrix 17(1):17–28. https://doi.org/10.4404/hystrix-17.1-4361
Ficetola GF, Pansu J, Bonin A, Coissac E, Giguet-Covex C, De Barba M, Gielly L, Lopes CM, Boyer F, Pompanon F, Rayé G, Taberlet P (2015) Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol Ecol Resour 15(3):543–556. https://doi.org/10.1111/1755-0998.12338
Fratini S, Zaccara S, Barbaresi S, Grandjean F (2005) Phylogeography of the threatened crayfish (genus Austropotamobius) in Italy: implications for its taxonomy and conservation. Heredity 94(1):108–118. https://doi.org/10.1038/sj.hdy.6800581
Füreder L, Gherardi F, Holdich D, Reynolds J, Sibley P, Souty-Grosset C (2010) In: IUCN 2010. The IUCN Red List of Threatened Species 2010.3: e.T2430A9438817
Gaudiano L, Cascella A, L’Erario P, Corriero G (2023) First data on the presence of Lutra lutra in Bosco Incoronata Natural Regional Park (Apulia, South Italy). BORNH Bull Reg Nat Hist 3(1):1–11. https://doi.org/10.6093/2724-4393/9985
Holdich D (2003) Ecology of the White-clawed Crayfish. Conserving Natura 2000 Rivers Ecology Series No. 1. English Nature, Peterborough
IUCN (2023) The IUCN Red list of threatened species. Version 2022-2. https://www.iucnredlist.org
Jerde CL, Mahon AR, Chadderton WL, Lodge DM (2011) Sight-unseen detection of rare aquatic species using environmental DNA: eDNA surveillance of rare aquatic species. Conserv Lett 4(2):150–157. https://doi.org/10.1111/j.1755-263X.2010.00158.x
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, Jackson CA, Lance RF, Mangan AM, Monroe EM, Piaggio AJ, Stokdyk JP, Wilson CC, Rithcer CA (2020a) Reporting the limits of detection and quantification for environmental DNA assays. Environ DNA 2:271–282. https://doi.org/10.1002/edn3.29
Klymus KE, Ruiz Ramos DV, Thompson NL, Richter CA (2020b) Development and testing of species-specific quantitative PCR assays for environmental DNA applications. J Vis Exp 165:e61825. https://doi.org/10.3791/61825
Loy A (2018) Eurasian otter. Duplaix N. Savage M. The global Otter Conservation Strategy. IUCN/SSC Otter Specialist Group, Salem, Oregon, pp 46–56
Loy A, Kranz A, Oleynikov A, Roos A, Savage M, Duplaix N (2020) Lutra lutra. In IUCN 2020. The IUCN Red List of Threatened Species 2022: e.T12419A218069689
Malthieux L (2020) La Loutre d’Europe Lutra lutra (Linnaeus, 1758) en Roya- Bévéra: relique ou retour ? Prospections, état des lieux et implications. Faune-PACA Publication 98:22
Maxwell D, Jennings S (2005) Power of monitoring programmes to detect decline and recovery of rare and vulnerable fish. J Appl Ecol 42(1):25–37. https://doi.org/10.1111/j.1365-2664.2005.01000.x
Nadai GD, Cassol M, Lapini L (2022) First data on the natural recovery of the Eurasian otter (Lutra lutra Linnaeus, 1758) in Veneto Region (north-eastern Italy). Habitat Online. https://www.habitatonline.eu/2022/11/first-data-on-the-natural-recovery-of-the-eurasian-otter-lutra-l-lutra-linnaeus-1758-in-veneto-region-north-eastern-italy/
Pascher K, Švara V, Jungmeier M (2022) Environmental DNA-based methods in biodiversity monitoring of protected areas: application range, limitations, and needs. Diversity 14(6):463. https://doi.org/10.3390/d14060463
Pavanello M, Lapini L, Kranz A, Iordan F (2015) Rediscovering the eurasian otter (Lutra lutra L.) in Friuli Venezia Giulia and notes on its possible expansion in northern Italy. IUCN Otter Specialist Group Bull 32(1):12–20
Pawlowski J, Apothéloz-Perret-Gentil L, Mächler E, Altermatt F (2020) Environmental DNA applications for biomonitoring and bioassessment in aquatic ecosystems. Guidelines. Bern, Switzerland, Federal Office for the Environment. https://doi.org/10.5167/UZH-187800
Peralta D, Vaz-Freire T, Ferreira C, Mendes T, Mira A, Santos S, Alves PC, Lambin X, Beja P, Paupério J, Pita R (2023) From species detection to population size indexing: the use of sign surveys for monitoring a rare and otherwise elusive small mammal. Eur J Wildl Res 69:9. https://doi.org/10.1007/s10344-022-01634-2
Prigioni C, Balestrieri A, Remonti L (2007) Decline and recovery in otter Lutra lutra populations in Italy. Mammal Rev 37(1):71–79. https://doi.org/10.1111/j.1365-2907.2007.00105.x
Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E (2011) ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res 39(21):e145. https://doi.org/10.1093/nar/gkr732
Righetti D (2011) Return of the otter in South Tyrol (NE Italy). In: Prigioni C, Loy A, Balestrieri A, Remonti L (eds) Proceedings of the IUCN XI International Otter Colloquium. Hystrix It J Mamm Supp. https://doi.org/10.4404/hystrix-22.0-4744
Salvidio S, Mori M, Lattes A, Galli L, Arillo A (2002) The freshwater crayfish Austropotamobius pallipes (Lereboullet, 1858) in Liguria, NW Italy: implications for management at the regional level. Bull Fr Pêche Piscic 367:663–670. https://doi.org/10.1051/kmae:2002057
Sanz N, Franch N, Araguas RM, Viñas J, Vidal O (2023) Environmental DNA assay for the detection of the American Bullfrog (Lithobates catesbeianus) in the early stages of the Invasion in the Ebre Delta. Animals 13(4):683. https://doi.org/10.3390/ani13040683
Taberlet P, Griffin S, Goossens B, Questiau S, Manceau V, Escaravage N, Waits LP, Bouvet J (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 24(16):3189–3194. https://doi.org/10.1093/nar/24.16.3189
Taberlet P, Bonin A, Zinger L, Coissac E (2018) Environmental DNA: for Biodiversity Research and Monitoring. Oxford University Press, Oxford
Thomsen PF, Kielgast J, Iversen LL, Wiuf c, Rasmussen M, Gilbert MT, Orlando L, Willerslev E (2012) Monitoring endangered freshwater biodiversity using environmental DNA: species monitoring by environmental DNA. Mol Ecol 21(11):2565–2573. https://doi.org/10.1111/j.1365-294X.2011.05418.x
Troth CR, Burian A, Mauvisseau Q, Bulling M, Nigthingale J, Mauvisseau C, Sweet MJ (2020) Development and application of eDNA-based tools for the conservation of white-clawed crayfish. Sci Total Environ 748:141394. https://doi.org/10.1016/j.scitotenv.2020.141394
Tsuji S, Takahara T, Doi H, Shibata N, Yamanaka H (2019) The detection of aquatic macroorganisms using environmental DNA analysis - A review of methods for collection, extraction, and detection. Environ DNA 1:99–108. https://doi.org/10.1002/edn3.21
Vigna Taglianti A, Bologna MA (1982) La fauna. AA.VV. Piano per Il Parco Naturale Regionale Delle Alpi Liguri. Sistemi I, II, III, Regione Liguria
Williams-Subiza EA, Epele LB (2021) Drivers of biodiversity loss in freshwater environments: a bibliometric analysis of the recent literature. Aquat Conserv: Mar Freshw Ecosyst 31(9):2469–2480. https://doi.org/10.1002/aqc.3627
Xia Z, Johansson ML, Gao Y, Zhan L, Haffner GD, MacIsaac, Zhan A (2018) Conventional versus real-time quantitative PCR for rare species detection. Ecol Evol 8(23):11799–11807. https://doi.org/10.1002/ece3.4636
Funding
This work was supported by the Liguria Regional Authority (Italy) through grant number G55J19000450007 of the Biodiv’Connect programme of the European Interreg Biodiv’ALP France-Italy ALCOTRA 2014–2020 project. The authors acknowledge the support by the Italian Ministry of University and Research through the National Biodiversity Future Center (NBFC), part of the National Recovery and Resilience Plan, Mission 4, Component 2, Investment 1.4, Project CN00000033. The authors are also grateful to Stefano Cannicci for his useful comments to the manuscript.
Open access funding provided by Università degli Studi di Firenze within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Contributions
L.B., S.F. and A.I. wrote the manuscript. S.F, C.C., D.O., V.R., and A.I. conceptualized and designed the study. L.B., L.T. and G.C. collected the samples. L.B., C.N., A.I and S.F. performed lab work and provided lab support. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ballini, L., Ottonello, D., Repetto, V. et al. Early detection of rare and elusive endangered species using environmental DNA: a case study for the Eurasian otter and the white-clawed crayfish in northwestern Italy. Conserv Genet (2024). https://doi.org/10.1007/s10592-024-01619-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10592-024-01619-5