Abstract
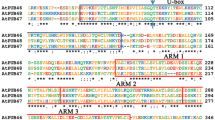
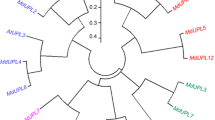
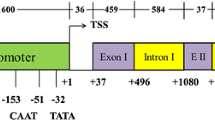
Ubiquitin-conjugating (UBC) enzyme is a key enzyme in ubiquitination. Here, we describe the cloning, characterization, and expression pattern of a novel gene, CmUBC, from a melon. Comparison of the deduced amino acid sequences allowed the identification of highly conserved motifs. Synteny analysis between Cucumis sativus L. and Arabidopsis demonstrated that homologs of several Cucumis UBC genes were found in corresponding syntenic blocks of Arabidopsis. The homology structure model of the CmUBC protein was constructed. UBCs from melon, yeast, and Arabidopsis were highly conserved in their three-dimensional folding. CmUBC was ubiquitously expressed in all melon tissues. Increased transcript levels of CmUBC were observed during drought and salinity stresses, which suggested that the expression of the CmUBC gene in melon plants is responsive to physiological water stress. These results suggested that the CmUBC gene might play an important role in the modulation of the ubiquitination pathway.







Similar content being viewed by others
References
Azad AK, Sato R, Ohtani K, Sawa Y, Ishikawa T, Shibata H (2011) Functional characterization and hyperosmotic regulation of aquaporin in Synechocystis sp. PCC 6803. Plant Sci 180:375–382
Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F (2001) Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci 6:463–470
Baloglu MC, Negre-Zakharov F, Öktem HA, Yücel M (2011) Molecular cloning, characterization and expression analysis of a gene encoding a Ran binding protein (RanBP) in Cucumis melo L. Turk J Biol 35:387–397
Baloglu MC, Öz MT, Öktem HA, Yücel M (2012) Expression analysis of TaNAC69-1 and TtNAMB-2, wheat NAC family transcription factor genes under abiotic stress conditions in durum wheat (Triticum turgidum). Plant Mol Biol Rep 30(5):1246–1252
Bhatnagar-Mathur P, Vadez V, Sharma KK (2008) Transgenic approaches for abiotic stress tolerance in plants: retrospect and prospects. Plant Cell Rep 27:411–424
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Cheng F, Wu J, Fang L, Wang X (2012) Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Plant Sci 3:198
Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101:15289–15294
Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15(3):532–534
Cook WJ, Jeffrey LC, Sullivan ML, Vierstra RD (1992) Three-dimensional structure of a ubiquitin-conjugating enzyme (E2). J Biol Chem 267(21):15116–15121
Cook WJ, Jeffrey LC, Xu Y, Chau V (1993) Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4. Biochemistry 32(50):13809–13817
Devoto A, Muskett PR, Shirasu K (2003) Role of ubiquitination in the regulation of plant defense against pathogens. Curr Opin Plant Biol 6:307–311
Goebl MG, Yochem J, Jentsch S, McGrath JP, Varshavsky A, Byers B (1988) The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 241(4871):1331–1335
Haas AL, Reback PB, Chau OV (1991) Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. J Biol Chem 266(8):5104–5112
Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS (2001) Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9(10):897–904
Han YH, Zhang ZH, Liu JH, Lu JY, Huang SW, Jin WW (2008) Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet Genome Res 122(1):80–88
Hellmann H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297:793–797
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1–32
Jiang YH, Beaudet AL (2004) Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr 16:419–426
Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139:1597–1611
Lee H, Xiong L, Gong Z (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev 15:912–924
Maiti R, Van Domselaar GH, Zhang H, Wishart DS (2004) SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res 32:590–594
Mukoko BJ, Vandeputte OM, Himanen K, Mol A, Vaessen Q, El Jaziri M, Baucher M (2010) Ectopic expression of PtaRHE1, encoding a poplar RING-H2 protein with E3 ligase activity, alters plant development and induces defencerelated responses. J Exp Bot 61:297–310
Paterson A, Wang X, Tang H, Lee T (2012) Synteny and genomic rearrangements. In: Wendel J, Greilhuber J, Dolezel J, Leitch IJ (eds) Plant genome diversity, vol 1. Springer, Vienna, pp 195–207
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Ren Y, Zhang Z, Liu J, Staub JE, Han Y et al (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS One 4:e5795
Reynolds P, Weber S, Prakash L (1985) RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci USA 82:168–172
Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng XW (2002) Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev 16:554–559
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang H, Wang X, Bowers JE, Ming R, Alam M et al (2008) Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 18:1944–1954
Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M (2010) A practical approach to RT-qPCR: publishing data that conform to the MIQE guidelines. Methods 50:S1–S5
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Van Nocker S, Vierstra RD (1991) Cloning and characterization of a 20-kDa ubiquitin carrier protein from wheat that catalyzes multiubiquitin chain formation in vitro (conjugation/protein degradation/ubiquitin mutant). Proc Natl Acad Sci USA 88:10297–10301
Wan X, Mo A, Liu S, Yang L, Li L (2011) Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression. J Biosci Bioeng 111(4):478–484
Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2(3):169–178
Worthylake DK, Prakash S, Prakash L, Hill CP (1998) Crystal structure of the Saccharomyces cerevisiae ubiquitin-conjugating enzyme Rad6 at 2.6 A resolution. J Biol Chem 273(11):6271–6276
Xiao W, Lin SL, Broomfield S, Chow BL, Wei YF (1998) The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved UBC-like protein family. Nucleic Acids Res 26(17):3908–3914
Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419:167–170
Xu L, Menard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH (2009) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 57:279–288
Yan J, Wang J, Li Q, Hwang JR, Patterson C, Zhang H (2003) AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol 132:861–869
Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19:1912–1929
Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UBCH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102(4):533–539
Zhou GA, Chang RZ, Qiu LJ (2010) Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol Biol 72:357–367
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zwirn P, Stary S, Luschnig C, Bachmair A (1997) Arabidopsis thaliana RAD6 homolog AtUBC2 complements UV sensitivity, but not N-end rule degradation deficiency, of Saccharomyces cerevisiae rad6 mutants. Curr Genet 32:309–314
Acknowledgments
This study was supported by the Research Fund of Kastamonu University Grant No. KÜBAP-01/2012-34.
Author information
Authors and Affiliations
Corresponding author
Additional information
GenBank Accession No. The CmUBC gene was deposited in GenBank under the accession number EU853458.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Comparison of Ub1 and Ub5 of melon CmUBC and yeast UBC4. (a) The 3D model of CmUBC (red) was first constructed with the experimental X-ray crystal structure of UBC4 (green) and then superposed on it. The superposed residues that form ubiquitin thioester intermediate interaction residues Ub1 (b) and Ub5 (c) in the superposed structures are shown as sticks. The residues of CmUBC are shown in red and those of the superposed structures in green. The RMSD value between the template UBC4 and the model CmUBC was 0.89 Å (JPEG 156 kb)
Supplementary Fig. 2
Comparison of Ub1, Ub4, Ub5, and E3 of melon CmUBC and Arabidopsis UBC1. (a) The 3D model of CmUBC (red) was first constructed with the experimental X-ray crystal structure of UBC1 (green) and then superposed on it. The superposed residues that form ubiquitin thioester intermediate interaction residues Ub1 (b), Ub4 (c), Ub5 (d), and E3 interaction residues (e) in the superposed structures are shown as sticks. The residues of CmUBC are shown in red and those of the superposed structures in green. The RMSD value between the template UBC1 and the model CmUBC was 1.71 Å (JPEG 208 kb)
Supplementary Fig. 3
Construction of standard curve for CmUBC gene quantification. Amplification curve of different concentrations of a pCR8/GW/TOPO-CmUBC plasmid DNA and its standard curve. Plasmid dilutions, left to right: (p7) 107, (p6) 106, (p5) 105, (p4) 104, (p3) 103, (p2) 102, and (p1) 101 copies/µL (TIFF 1375 kb)
Rights and permissions
About this article
Cite this article
Baloglu, M.C., Patir, M.G. Molecular Characterization, 3D Model Analysis, and Expression Pattern of the CmUBC Gene Encoding the Melon Ubiquitin-Conjugating Enzyme Under Drought and Salt Stress Conditions. Biochem Genet 52, 90–105 (2014). https://doi.org/10.1007/s10528-013-9630-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-013-9630-9