Abstract
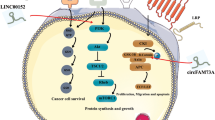
SNHG4 is a lncRNA that was previously reported to promote colorectal cancer (CRC) progression via molecular sponge mechanism. Bioinformatic analysis suggested SNHG4 might scaffold TAF15 protein-RNF14 mRNA interaction. We aimed to investigate the mechanisms of potential SNHG4/TAF15/RNF14 axis in promoting CRC malignant phenotypes. Protein-RNA interaction was determined using RNA immunoprecipitation, pull-down and fluorescence in situ hybridization (FISH) combined immunofluorescence assays. Cell apoptosis rates were quantified using flow cytometry. CCK-8 and colony formation were adopted to determine cell proliferation. Wound healing and transwell assays were employed to assess cell migration and invasion, respectively. Xenograft tumor model was applied to assess the effects of SNHG4 on CRC tumorigenesis in vivo. SNHG4, TAF15 and RNF14 were up-regulated in CRC tissues. SNHG4 overexpression promoted cell proliferation, migration, invasion, and Wnt/β-catenin pathway activation in vitro, as well as tumor growth in vivo. The inhibited malignant phenotypes caused by SNHG4 knockdown were impeded by TAF15 or RNF14 overexpression. Mechanistically, SNHG4 recruited TAF15 protein and thus promoted the interaction between TAF15 protein and RNF14 mRNA, leading to the increased RNF14 mRNA stability. This in turn facilitated the Wnt/β-catenin signal transduction. SNHG4 enhanced RNF14 mRNA stability and activated the Wnt/β-catenin pathway to promote the progression of colorectal cancer by recruiting TAF15 protein.









Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CRC:
-
Colorectal cancer
- FISH:
-
Fluorescence in situ hybridization
- lncRNAs:
-
Long non-coding RNAs
- EMT:
-
Epithelial-mesenchymal transition
- SNHG4:
-
Small nucleolar RNA host gene 4
- TAF15:
-
TATA-box binding protein associated factor 15
- TCF/LEF:
-
T-cell factor/lymphoid enhancer-binding factor
- RNF:
-
Ring finger
- DMEM:
-
Dulbecco’s Modified Eagle’s Medium
- RL:
-
Right lower
- RU:
-
Right upper
References
Chen W et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132
Cao W, Chen HD, Yu YW, Li N, Chen WQ (2021) Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 134:783–791
Vu T, Datta PK (2017) Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers (Basel) 9:171
Pavlič A, Urh K, Štajer K, Boštjančič E, Zidar N (2021) Epithelial-Mesenchymal transition in colorectal carcinoma: comparison between primary tumor, lymph node and liver metastases. Front Oncol 11:1504
Sugai T et al (2018) Analysis of the expression of cancer-associated fibroblast- and EMT-related proteins in submucosal invasive colorectal cancer. J Cancer 9:2702–2712
Zhang Z et al (2020) Genomics and prognosis analysis of epithelial-mesenchymal transition in colorectal cancer patients. BMC Cancer 20:1–2
Yang Y et al (2021) Comprehensive analysis of EMT-related genes and lncRNAs in the prognosis, immunity, and drug treatment of colorectal cancer. J Transl Med 19:1–21
Mo S et al (2021) Comprehensive transcriptomic analysis reveals prognostic value of an EMT-related gene signature in colorectal cancer. Front cell Dev Biol 9:1566
Iyer MK et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208
Fang Y, Fullwood MJ (2016) Roles, functions, and mechanisms of long non-CODING RNAs in cancer. Genomics Proteomics Bioinform 14:42–54
Yao RW, Wang Y, Chen LL (2019) Cellular functions of long noncoding RNAs. Nat Cell Biol 21:542–551
Peng WX, Koirala P, Mo YY (2017) LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36:5661–5667
Chi Y, Wang D, Wang J, Yu W, Yang J (2019) Long non-coding RNA in the Pathogenesis of Cancers. Cells 8:1015
Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21:22–36
Jiao Y et al (2020) The prognostic value of lncRNA SNHG4 and its potential mechanism in liver cancer. Biosci Rep. https://doi.org/10.1042/BSR20190729
Cheng XB et al (2021) Knockdown of lncRNA SNHG4 suppresses gastric cancer cell proliferation and metastasis by targeting miR-204-5p. Neoplasma 68:546–556
Wang S, Zhu W, Qiu J, Chen F (2021) lncRNA SNHG4 promotes cell proliferation, migration, invasion and the epithelial-mesenchymal transition process via sponging miR-204–5p in gastric cancer. Mol Med Rep 23:1–11
Zhou Z et al (2021) lncRNA SNHG4 modulates colorectal cancer cell cycle and cell proliferation through regulating miR-590–3p/CDK1 axis. Aging (Albany. NY). 13:9838–9858
Wang X et al (2021) LncRNA SNHG14 promotes cell proliferation and invasion in colorectal cancer through modulating miR-519b-3p/DDX5 axis. J Cancer 12:4958–4970
Kc W, Hy C (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914
Andric V et al (2021) A scaffold lncRNA shapes the mitosis to meiosis switch. Nat Commun 12:1–2
Bertolotti A, Lutz Y, Heard DJ, Chambon P, Tora L (1996) hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J 15:5022
Ruan X et al (2020) lncRNA LINC00665 Stabilized by TAF15 Impeded the Malignant Biological Behaviors of Glioma Cells via STAU1-Mediated mRNA Degradation. Mol Ther Nucleic Acids 20:823–840
Ibrahim F et al (2013) Identification of in vivo, conserved, TAF15 RNA binding sites reveals the impact of TAF15 on the neuronal transcriptome. Cell Rep 3:301–308
Ren P, Xing L, Hong X, Chang L, Zhang H (2020) LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. Cancer Med 9:7706–7716
Pan Li, Li Yi, Jin Li, Li J, Aixiang Xu (2020) TRPM2-AS promotes cancer cell proliferation through control of TAF15. Int J Biochem Cell Biol 120:105683
Lin Y et al (2020) RNAInter in 2020: RNA interactome repository with increased coverage and annotation. Nucleic Acids Res 48:D189–D197
Schatoff EM, Leach BI, Dow LE (2017) Wnt signaling and colorectal cancer. Curr Colorectal Cancer Rep 13:101
Cheng X, Xu X, Chen D, Zhao F, Wang W (2019) Therapeutic potential of targeting the Wnt/β-catenin signaling pathway in colorectal cancer. Biomed Pharmacother 110:473–481
Sebio A, Kahn M, Lenz HJ (2014) The potential of targeting Wnt/β-catenin in colon cancer. Expert Opin Ther Targets 18:611–615
Zhao H et al (2022) Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol Cancer 21:1–34
Hao HX et al (2012) ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485:195–202
Thomas JJ et al (2016) RNF4-dependent oncogene activation by protein stabilization. Cell Rep 16:3388–3400
Wu B et al (2013) Ring finger protein 14 is a new regulator of TCF/β-catenin-mediated transcription and colon cancer cell survival. EMBO Rep 14:347–355
Chandrashekar DS et al (2017) UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19:649–658
Tang Z et al (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45:W98–W102
Tetsu O, McCormick F (1999) Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422–426
He TC et al (1998) Identification of c-MYC as a target of the APC pathway. Science 281:1509–1512
Wu B, Crampton SP, Hughes CCW (2007) Wnt signaling induces MMP expression and regulates T cell transmigration. Immunity 26:227
Jamora C, DasGupta R, Kocieniewski P, Fuchs E (2003) Links between signal transduction, transcription and adhesion in epithelial bud development. Nature 422:317–322
Loh C-Y et al (2019) The E-Cadherin and N-Cadherin Switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells 8:1118
Acknowledgements
Not applicable.
Funding
This work was supported by Guangxi Research Foundation for Science &Technology Base and Talent Special (no. AD20238005).
Author information
Authors and Affiliations
Contributions
LL: Conceptualization; Writing-original draft; Methodology; Formal analysis; Supervision; Validation; Visualization; BH: Data curation; Resources; LY: Investigation; Software; LZ: Funding acquisition; Project administration; Writing-review & editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
The approval for patient recruitment, specimen collection and tissue processing has been acquired from the clinical research ethics committee in Affiliated Hospital of Guilin Medical University. The animal research ethics committee of Affiliated Hospital of Guilin Medical University approved all the animal experiment procedures.
Consent for publication
The informed consent was obtained from study participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lv, L., Huang, B., Yi, L. et al. Long non-coding RNA SNHG4 enhances RNF14 mRNA stability to promote the progression of colorectal cancer by recruiting TAF15 protein. Apoptosis 28, 414–431 (2023). https://doi.org/10.1007/s10495-022-01781-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-022-01781-6