Abstract
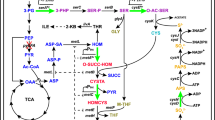
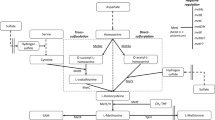
Relieving the feedback inhibition of key enzymes in a metabolic pathway is frequently the first step of producer-strain construction by genetic engineering. However, the strict feedback regulation exercised by microorganisms in methionine biosynthesis often makes it difficult to produce methionine at a high level. In this study, Corynebacterium glutamicum ATCC 13032 was metabolically engineered for methionine production. First, the metD gene encoding the methionine uptake system was deleted to achieve extracellular accumulation of methionine. Then, random mutagenesis was performed to remove feedback inhibition by metabolic end-products. The resulting strain C. glutamicum ENM-16 was further engineered to block or decrease competitive branch pathways by deleting the thrB gene and changing the start codon of the dapA gene, followed by point mutations of lysC (C932T) and pyc (G1A, C1372T) to increase methionine precursor supply. To enrich the NADPH pool, glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase in the pentose phosphate pathway were mutated to reduce their sensitivity to inhibition by intracellular metabolites. The resultant strain C. glutamicum LY-5 produced 6.85 ± 0.23 g methionine l−1 with substrate-specific yield (Y P/S) of 0.08 mol per mol of glucose after 72 h fed-batch fermentation. The strategies described here will be useful for construction of methionine engineering strains.





Similar content being viewed by others
References
Antoine FR, Wei CI, Littell RC, Marshall MR (1999) HPLC method for analysis of free amino acids in fish using o-phthaldialdehyde precolumn derivatization. J Agric Food Chem 47(12):5100–5107
Becker J, Klopprogge C, Herold A, Zelder O, Bolten CJ, Wittmann C (2007) Metabolic flux engineering of l-lysine production in Corynebacterium glutamicum - over expression and modification of G6P dehydrogenase. J Biotechnol 132(2):99–109. doi:10.1016/j.jbiotec.2007.05.026
Belfaiza J, Martel A, Margarita D, Saint Girons I (1998) Direct sulfhydrylation for methionine biosynthesis in Leptospira meyeri. J Bacteriol 180(2):250–255
Driouch H, Melzer G, Wittmann C (2012) Integration of in vivo and in silico metabolic fluxes for improvement of recombinant protein production. Metab Eng 14(1):47–58. doi:10.1016/j.ymben.2011.11.002
Hausmann R (2002) Molecular clones. Springer, The Netherlands. doi:10.1007/978-94-017-3540-7_20
Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes—gene-splicing by overlap extension. Gene 77(1):61–68. doi:10.1016/0378-1119(89)90359-4
Hwang BJ, Kim Y, Kim HB, Hwang HJ, Kim JH, Lee HS (1999) Analysis of Corynebacterium glutamicum methionine biosynthetic pathway: isolation and analysis of metB encoding cystathionine gamma-synthase. Mol Cells 9(3):300–308
Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertes AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiol-SGM 153(8):2491–2504. doi:10.1099/mic.0.2006/005587-0
Jakobsen OM, Brautaset T, Degnes KF, Heggeset TM, Balzer S, Flickinger MC, Valla S, Ellingsen TE (2009) Overexpression of wild-type aspartokinase increases l-lysine production in the thermotolerant methylotrophic bacterium Bacillus methanolicus. Appl Environ Microbiol 75(3):652–661. doi:10.1128/AEM.01176-08
Kase H, Nakayama K (1975) l-methionine production by methionine analog-resistant mutants of Corynebacterium glutamicum. Agric Biol Chem 39:153–160
Kim JW, Kim HJ, Kim Y, Lee MS, Lee HS (2001) Properties of the Corynebacterium glutamicum metC gene encoding cystathionine beta-lyase. Mol Cells 11(2):220–225
Kim SY, Lee J, Lee SY (2015) Metabolic engineering of Corynebacterium glutamicum for the production of L-ornithine. Biotechnol Bioeng 112(2):416–421. doi:10.1002/bit.25440
Kovaleva GY, Gelfand MS (2007) Regulation of methionine/cysteine biosynthesis in Corynebacterium glutamicum and related organisms. Mol Biol 41(1):126–136. doi:10.1134/S0026893307010177
Kroemer JO, Wittmann C, Schroeder H, Heinzle E (2006) Metabolic pathway analysis for rational design of l-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab Eng 8(4):353–369. doi:10.1016/j.ymben.2006.02.001
Kromer JO, Heinzle E, Schroder H, Wittmann C (2006) Accumulation of homolanthionine and activation of a novel pathway for isoleucine biosynthesis in Corynebacterium glutamicum McbR deletion strains. J Bacteriol 188(2):609–618. doi:10.1128/JB.188.2.609-618.2006
Kumar D, Gomes J (2005) Methionine production by fermentation. Biotechnol Adv 23(1):41–61. doi:10.1016/j.biotechadv.2004.08.005
Kumar D, Garg S, Bisaria VS, Sreekrishnan TR, Gomes J (2003) Production of methionine by a multi-analogue resistant mutant of Corynebacterium lilium. Process Biochem 38(8):1165–1171. doi:10.1016/S0032-9592(02)00287-X
March JC, Eiteman MA, Altman E (2002) Expression of an anaplerotic enzyme, pyruvate carboxylase, improves recombinant protein production in Escherichia coli. Appl Environ Microbiol 68(11):5620–5624
Meiswinkel TM, Gopinath V, Lindner SN, Nampoothiri KM, Wendisch VF (2013) Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb Biotechnol 6(2):131–140
Ohnishi J, Katahira R, Mitsuhashi S, Kakita S, Ikeda M (2005) A novel gnd mutation leading to increased l-lysine production in Corynebacterium glutamicum. FEMS Microbiol Lett 242(2):265–274. doi:10.1016/j.femsle.2004.11.014
Park S, Lee J, Sim S, Kim Y, Lee H (2007) Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metab Eng 9(4):327–336. doi:10.1016/j.ymben.2007.05.001
Park SH, Kim HU, Kim TY, Park JS, Kim S, Lee SY (2014) Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat Commun 5:4618. doi:10.1038/ncomms5618
Rey DA, Puhler A, Kalinowski J (2003) The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J Bacteriol 103(1):51–65. doi:10.1016/S0168-1656(03)00073-7
Rey DA, Nentwich SS, Koch DJ, Ruckert C, Puhler A, Tauch A, Kalinowski J (2005) The McbR repressor modulated by the effector substance S-adenosylhomocysteine controls directly the transcription of a regulon involved in sulphur metabolism of Corynebacterium glutamicum ATCC 13032. Mol Microbiol 56(4):871–887. doi:10.1111/j.1365-2958.2005.04586.x
Rueckert C, Milse J, Albersmeier A, Koch DJ, Puehler A, Kalinowski J (2008) The dual transcriptional regulator CysR in Corynebacterium glutamicum ATCC 13032 controls a subset of genes of the McbR regulon in response to the availability of sulphide acceptor molecules. BMC Genom 9:483. doi:10.1186/1471-2164-9-483
Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145(1):69–73
Shen T, Shen W, Xiong Y, Liu H, Zheng H, Zhou H, Rui B, Liu J, Wu J, Shi Y (2009) Increasing the accuracy of mass isotopomer analysis through calibration curves constructed using biologically synthesized compounds. J Mass Spectrom 44(7):1066–1080. doi:10.1002/jms.1583
Tauch A, Kirchner O, Loffler B, Gotker S, Puhler A, Kalinowski J (2002) Efficient electrotransformation of Corynebacterium diphtheriae with a mini-replicon derived from the Corynebacterium glutamicum plasmid pGA1. Curr Microbiol 45(5):362–367. doi:10.1007/s00284-002-3728-3
Tosaka O, Takinami K (1986) Biotechnology of amino acid production. In: Aida K, Chibata I, Nakayama K (eds) Progress in industrial microbiology, vol 24, 24th edn. Elsevier, Tokyo, pp 152–172
Trotschel C, Deutenberg D, Bathe B, Burkovski A, Kramer R (2005) Characterization of methionine export in Corynebacterium glutamicum. J Bacteriol 187(11):3786–3794. doi:10.1128/JB.187.11.3786-3794.2005
Vogt M, Haas S, Klaffl S, Polen T, Eggeling L, van Ooyen J, Bott M (2014) Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for l-leucine overproduction. Metab Eng 22:40–52. doi:10.1016/j.ymben.2013.12.001
Willke T (2014) Methionine production—a critical review. Appl Microbiol Biotechnol 98(24):9893–9914. doi:10.1007/s00253-014-6156-y
Xu J, Han M, Zhang J, Guo Y, Zhang W (2014) Metabolic engineering Corynebacterium glutamicum for the l-lysine production by increasing the flux into l-lysine biosynthetic pathway. Amino Acids 46(9):2165–2175. doi:10.1007/s00726-014-1768-1
Acknowledgments
This research was supported by Twelfth Five-Year Plan National Science and Technology Program on Rural Area (2014BAL02B00). We are grateful to Professor Jihui Wang (Dalian Polytechnic University, China) for the kind help on the experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Cong, H., Liu, B. et al. Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level. Antonie van Leeuwenhoek 109, 1185–1197 (2016). https://doi.org/10.1007/s10482-016-0719-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0719-0