Abstract
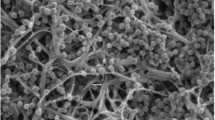
An actinomycete strain SL3-70T was isolated from a rice field and characterised using a polyphasic approach. The morphological and chemotaxonomical characteristics of strain SL3-70T indicate that it belongs to the genus Micromonospora. The phylogenetic analysis of the nearly complete 16S rRNA gene sequence revealed that strain SL3-70T is a member of the genus Micromonospora, and is closely related to Micromonospora echinaurantica DSM 43904T (99.1 % 16S rRNA gene sequence similarity) and Micromonospora kangleipakensis MBRL 34T (98.8 %). DNA–DNA relatedness between strain SL3-70T and its relatives ranged from 21.2 % ± 0.6 to 38.7 % ± 0.4. The results obtained from our study indicate that strain SL3-70T represents a novel species of the genus Micromonospora, for which the name Micromonospora soli sp. nov. is proposed. The type strain is SL3-70T (=BCC 67268T; =NBRC 110009T).



Similar content being viewed by others
References
Arai T (1975) Culture media for actinomycetes. The Society for Actinomycetes, Tokyo
Bérdy J (2005) Bioactive microbial metabolites. J Antibiot (Tokyo) 58:1–26
Carro L, Pukall R, Spröer C, Kroppenstedt RM, Trujillo ME (2012) Micromonospora cremea sp. nov. and Micromonospora zamorensis sp. nov., isolated from the rhizosphere of Pisum sativum. Int J Syst Evol Microbiol 62:2971–2977
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Euzéby JP (2010) List of bacterial names with standing in nomenclature. http://www.bacterio.cict.fr/
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid- deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Fang B, Liu C, Guan X, Song J, Zhao J, Liu H, Li C, Ning W, Wang X, Xiang W (2015) Two new species of the genus Micromonospora: Micromonospora palomenae sp. nov. and Micromonospora harpali sp. nov. isolated from the insects. Antonie Van Leeuwenhoek 108:141–150
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Garcia LC, Martínez-Molina E, Trujillo ME (2010) Micromonospora pisi sp. nov., isolated from root nodules of Pisum sativum. Int J Syst Evol Microbiol 60:331–337
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Itoh T, Kudo T, Parenti F, Seino A (1989) Amended description of the genus Kineosporia, based on chemotaxonomic and morphological studies. Int J Syst Bacteriol 39:168–173
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kawamoto I (1989) Genus Micromonospora. In: Williams ST, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 4. Williams & Wilkins, Baltimore, pp 2442–2450
Kawamoto I, Oka T, Nara T (1981) Cell wall composition of Micromonospora olivasterospora, Micromonospora sagamiensis, and related organisms. J Bacteriol 146:527–534
Kelly KL (1964) Inter-Society Color Council—National Bureau of Standard Color Name Charts Illustrated with Centroid Colors published in US
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kroppenstedt RM (1985) Fatty acid and menaquinone analysis of actinomycetes and related organisms. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 173–199
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Li L, Tang YL, Wei B, Xie QY, Deng Z, Hong K (2013) Micromonospora sonneratiae sp. nov., isolated from a root of Sonneratia apetala. Int J Syst Evol Microbiol 63:2383–2388
Li C, Liu CX, Zhao JW, Zhang YJ, Gao RX, Zhang XH, Yao M, Wang XJ, Xiang WS (2014) Micromonospora maoerensis sp. nov., isolated from a Chinese pine forest soil. Antonie Van Leeuwenhoek 105:451–459
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nakajima Y, Kitpreechavanich V, Suzuki K, Kudo T (1999) Microbispora corallina sp. nov., a new species of the genus Microbispora isolated from Thai soil. Int J Syst Bacteriol 49:1761–1767
Ørskov J (1923) Investigations into the morphology of the ray fungi. Levin and Munksgaard, Copenhagen
Ren J, Li L, Wei B, Tang YL, Deng ZX, Sun M, Hong K (2013) Micromonospora wenchangensis sp. nov., isolated from mangrove soil. Int J Syst Evol Microbiol 63:2389–2395
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101 published in US
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington DC, pp 607–654
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamaoka J (1994) Determination of DNA base composition. In: Goodfellow M, O’Donnell AG (eds) Chemical methods in prokaryotic systematics. Wiley, Chichester, pp 463–470
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Thawai C (2015) Micromonospora costi sp. nov., isolated from a leaf of Costus speciosus. Int J Syst Evol Microbiol 65:1456–1461
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uchida K, Aida K (1984) An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J Gen Appl Microbiol 30:131–134
Vela Gurovic MS, Müller S, Domin N, Seccareccia I, Nietzsche S, Martin K, Nett M (2013) Micromonospora schwarzwaldensis sp. nov., a producer of telomycin, isolated from soil. Int J Syst Evol Microbiol 63:3812–3817
Wang C, Xu XX, Qu Z, Wang HL, Lin HP, Xie QY, Ruan JS, Hong K (2011) Micromonospora rhizosphaerae sp. nov., isolated from mangrove rhizosphere soil. Int J Syst Evol Microbiol 61:320–324
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Williams ST, Cross T (1971) Actinomycetes. In: Booth C (ed) Methods in microbiology, vol 4. Academic Press, London, pp 295–334
Xiang W, Yu C, Liu C, Zhao J, Yang L, Xie B, Li L, Hong K, Wang X (2014) Micromonospora polyrhachis sp. nov., an actinomycete isolated from edible Chinese black ant (Polyrachis vicina Roger). Int J Syst Evol Microbiol 64:495–500
Xu P, Li WJ, Tang SK, Zhang TQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Zhao J, Guo L, He H, Liu C, Zhang Y, Li C, Wang X, Xiang W (2014) Micromospora taraxaci sp. nov., a novel endophytic actinomycete isolated from dandelion root (Taraxamcum mongloicum Hand.-Mazz.). Antonie Van Leeuwenhoek 106:667–674
Acknowledgments
We express our gratitude to Prof. Aharon Oren and Prof. Bernhard Schink for their valuable suggestion on naming bacterial species. This research was financially supported by the King Mongkut’s Institute of Technology Research Fund (KReF) and partially supported by Department of Biology and Actinobacterial Research Unit, Faculty of Science, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession numbers for the partial sequences of the 16S rRNA and gyrB genes of strain SL3-70T are AB981051 and LC030204, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thawai, C., Kittiwongwattana, C., Thanaboripat, D. et al. Micromonospora soli sp. nov., isolated from rice rhizosphere soil. Antonie van Leeuwenhoek 109, 449–456 (2016). https://doi.org/10.1007/s10482-016-0651-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-016-0651-3