Abstract
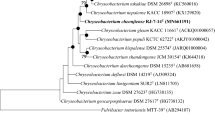
A novel strain, DCY107T, was isolated from soil collected from a ginseng field in Gochang, Republic of Korea. Strain DCY107T is Gram-negative, yellow pigmented, non-motile, non-flagellate, rod-shaped and aerobic. The strain was found to grow optimally at 25–30 °C and pH 6.5–7. Phylogenetically, strain DCY107T is closely related to Chryseobacterium polytrichastri DSM 26899T (98.49 % 16S rRNA gene sequence similarity), Chryseobacterium yeoncheonense JCM 18516T (97.78 %), Chryseobacterium aahli LMG 27338T (97.74 %), Chryseobacterium limigenitum LMG28734T (97.74 %), Chryseobacterium ginsenosidimutans JCM 16719T (97.47 %) and Chryseobacterium gregarium LMG 24052T (97.31 %). The DNA–DNA relatedness values between strain DCY107T and reference strains were found to be clearly below 70 %. The DNA G+C content of strain DCY107T was determined to be 34.2 mol%. The predominant quinone was identified menaquinone 6 (MK-6). The major polar lipids were identified as phosphatidylethanolamine and unidentified lipids: aminolipids AL1, AL2 and lipid L2. C16:00, iso-C15:00, iso-C15:02OH, iso-C17:03OH and summed feature 9 (iso-C17:1 ω9c and/or C16:0 10-methyl) were identified as the major fatty acids present in strain DCY107T. The results of physiological and biochemical tests allowed strain DCY107T to be differentiated phenotypically from other recognised species belonging to the genus Chryseobacterium. Therefore, it is suggested that the newly isolated organism represents a novel species, for which the name Chryseobacterium panacis sp. nov. is proposed, with the type strain designated as DCY107T (=CCTCC AB 2015195T = KCTC 42750T).

Similar content being viewed by others
References
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the Pseudomonas based on 16S rRNA sequence. Int J Syst Evol Microbiol 50:1563–1589
Bauer AW, Kirby WM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Path 45:493–496
Behrendt U, Ulrich A, Schumann P (2008) Chryseobacterium gregarium sp. nov., isolated from decaying plant material. Int J Syst Evol Microbiol 58:1069–1074
Bernardet JF, Nakagawa Y, Holmes B (2002) Subcommittee on the taxonomy of Flavobacterium & Cytophaga-like bacteria of the International Committee on Systematics of Prokaryotes. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Bernardet JF, Bruun B, Hugo C (2006) The genera Chryseobacterium and Elizabethkingia. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes, vol 7. Springer, New York, pp 638–676
Chen XY, Zhao R, Chen ZL, Liu L, Li XD, Li YH (2015) Chryseobacterium polytrichastri sp. nov., isolated from a moss (Polytrichastrum formosum), and emended description of the genus Chryseobacterium. Antonie Van Leeuwenhoek 107(2):403–410
Christensen WB (1946) Urea decomposition as a means of differentiating proteus and paracolon cultures from each other and from Salmonella and Shigella types. J Bacteriol 52:461–466
Collins MD (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
De Beer H, Hugo CJ, Jooste PJ, Willems A, Vancanneyt M, Coenye T, Vandamme PAR (2005) Chryseobacterium vrystaatense sp. nov., isolated from raw chicken in a chicken-processing plant. Int J Syst Evol Microbiol 55:2149–2153
Du J, Ngo HT, Won K, Kim KY, Jin FX, Yi TH (2015) Chryseobacterium solani sp. nov. isolated from field-grown eggplant rhizosphere soil. Int J Syst Evol Microbiol 65(9):2949–2954
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Evol Microbiol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hanh VV, Thi QD, Rita G, Dung NN (2013) Effectiveness of antagonistic bacterial metabolites to control Rhizoctonia solani on lettuces and Fusarium oxysporum on tomatoes. Korean J Microbiol Biotechnol 41:70–78
Hantsis-Zacharov E, Shake DT, Senderovich Y, Halpern M (2008) Chryseobacterium oranimense sp. nov., a psychrotolerant, proteolytic and lipolytic bacterium isolated from raw cow’s milk. Int J Syst Evol Microbiol 58:2635–2639
Herzog P, Winkler I, Wolking D, Kampfer P, Lipski A (2008) Chryseobacterium ureilyticum sp. nov., Chryseobacterium gambrini sp. nov., Chryseobacterium pallidum sp. nov. and Chryseobacterium molle sp. nov., isolated from beer-bottling plants. Int J Syst Evol Microbiol 58:26–33
Hoang VA, Kim YJ, Nguyen NL, Yang DC (2013) Chryseobacterium yeoncheonense sp. nov., with ginsenoside converting activity isolated from soil of a ginseng field. Arch Microbiol 195:463–471
Holmes B, Steigerwalt AG, Nicholson ACA (2013) DNA–DNA hybridization study of strains of Chryseobacterium, Elizabethkingia and Empedobacter and of other usually indoleproducing non-fermenters of CDC groups IIc, IIe, IIh and IIi, mostly from human clinical sources, and proposals of Chryseobacterium bernardetii sp. nov., Chryseobacterium carnis sp. nov., Chryseobacterium lactis sp. nov., Chryseobacterium nakagawai sp. nov. and Chryseobacterium taklimakanense comb. nov. Int J Syst Evol Microbiol 63:4639–4662
Im WT, Yang JE, Kim SY, Yi TH (2011) Chryseobacterium ginsenosidimutans sp. nov., a bacterium with Ginsenoside-converting activity isolated from soil of a Rhus vernicifera-cultivated field. Int J Syst Evol Microbiol 61:1430–1435
Jiao X, Lu X, Chen AJ, Luo Y, Hao JJ, Gao W (2015) Effects of Fusarium solani and F. oxysporum Infection on the metabolism of ginsenosides in American ginseng roots. Molecules 20(6):10535–10552
Kämpfer P, Dreyer U, Neef A, Dott W, Busse HJ (2003) Chryseobacterium defluvii sp. nov., isolated from waste water. Int J Syst Evol Microbiol 53:93–97
Kämpfer P, Vaneechoutte M, Lodders N, De Baere T, Avesani V, Janssens M, Busse HJ, Wauters G (2009) Description of Chryseobacterium anthropi sp. nov. to accommodate clinical isolates biochemically similar to Kaistella koreensis and Chryseobacterium haifense, proposal to reclassify Kaistella koreensis as Chryseobacterium koreense comb. nov. and emended description of the genus Chryseobacterium. Int J Syst Evol Microbiol 59:2421–2428
Kämpfer P, Chandel K, Prasad GB, Shouche YS, Veer V (2010) Chryseobacterium culicis sp. nov., isolated from the midgut of the mosquito Culex quinquefasciatus. Int J Syst Evol Microbiol 60:2387–2391
Kämpfer P, Poppel MT, Wilharm G, Busse HJ, McInroy JA, Glaeser SP (2014) Chryseobacterium gallinarum sp. nov., isolated from a chicken, and Chryseobacterium contaminans sp. nov., isolated as a contaminant from a rhizosphere sample. Int J Syst Evol Microbiol 64:1419–1427
Kämpfer P, Busse HJ, McInroy JA, Glaeser SP (2015a) Chryseobacterium arachidiradicis sp. nov. isolated from the geocarposphere (soil around the peanut) of very immature peanuts (Arachis hypogaea). Int J Syst Evol Microbiol 65:2179–2186
Kämpfer P, Trček J, Skok B, Šorgo A, Glaeser SP (2015b) Chryseobacterium limigenitum sp. nov., isolated from dehydrated sludge. Antonie Van Leeuwenhoek 107(6):1633–1638
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge
Kirk KE, Hoffman JA, Smith KA, Strahan BL, Failor KC, Krebs JE, Gale AN, Do TD, Sontag TC (2013) Chryseobacterium angstadtii sp. nov., isolated from a newt tank. Int J Syst Evol Microbiol 63:4777–4783
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–176
Loch TP, Faisal M (2014) Chryseobacterium aahli sp. nov., isolated from lake trout (Salvelinus namaycush) and brown trout (Salmo trutta), and emended descriptions of Chryseobacterium ginsenosidimutans and Chryseobacterium gregarium. Int J Syst Evol Microbiol 64:1573–1579
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C Content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Odonnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Opelt K, Berg G (2004) Diversity and antagonistic potential of bacteria associated with bryophytes from nutrient-poor habitats of the baltic sea coast. Appl Environ Microbiol 70:6569–6579
Park MS, Jung SR, Lee KH, Lee MS, Do JO, Kim SB, Bae KS (2006) Chryseobacterium soldanellicola sp. nov. and Chryseobacterium taeanense sp. nov., isolated from roots of sand-dune plants. Int J Syst Evol Microbiol 56:433–438
Rahman M, Punja ZK (2005) Biochemistry of ginseng root tissues affected by rusty root symptoms. Plant Physiol Biochem 43(12):1103–1114
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sang MK, Kim HS, Myung IS, Ryu CM, Kim BS, Kim KD (2013) Chryseobacterium kwangjuense sp. nov., isolated from pepper (Capsicum annuum L.) root. Int J Syst Evol Microbiol 63:2835–2840
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Skerman VBD (1967) A Guide to the identification of the genera of bacteria, 2nd edn. Williams and Wilkins, Baltimore
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Vandamme P, Bernardet JF, Segers P, Kersters K, Holmes B (1994) New perspectives in the classification of the Flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Bacteriol 44:827–831
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE (1987) Report of the Ad Hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wu YF, Wu QL, Liu SJ (2013) Chryseobacterium taihuense sp. nov., isolated from a eutrophic lake, and emended descriptions of the genus Chryseobacterium, Chryseobacterium taiwanense, Chryseobacterium jejuense and Chryseobacterium indoltheticum. Int J Syst Evol Microbiol 63:913–919
Yassin AF, Hupfer H, Siering C, Busse HJ (2010) Chryseobacterium treverense sp. nov., isolated from a human clinical source. Int J Syst Evol Microbiol 60:1993–1998
Acknowledgments
This research was supported by the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET NO: 313038-03-2-SB010) and also supported by a grant from the Next-Generation BioGreen 21 Program, Systems & Synthetic Agrobiotech Center (SSAC # PJ01116602), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2015_620_MOESM2_ESM.tif
Supplementary Fig. S2. Antagonistic activity of DCY107T and Chryseobacterium yeoncheonense JCM 18516T against Fusarium solani KACC 44891T. a Control, b C. yeoncheonense JCM 18516T, and c C. panacis KCTC 42750T. Supplementary material 2 (TIFF 590 kb)
10482_2015_620_MOESM3_ESM.tif
Supplementary Fig. S3. Maximum likelihood phylogenetic tree based on 16S rRNA gene sequences showing the position of strain DCY107T among species of the Chryseobacterium genus. Bootstrap values >50 % based on 1000 replications are shown at branching points. Scale bar, 0.01 substitutions per nucleotide position. Supplementary material 3 (TIFF 1720 kb)
10482_2015_620_MOESM4_ESM.tif
Supplementary Fig. S4. Two-dimensional TLC of total polar lipids. Strain DCY107T and Chryseobacterium yeoncheonense JCM 18516T were stained for total polar lipids with 5 % ethanolic molybdophosphoric acid. Abbreviations:PE phosphatidylethanolamine. a C. panacis KCTC 42750T; b Chryseobacterium yeoncheonense JCM 18516T. Supplementary material 4 (TIFF 736 kb)
Rights and permissions
About this article
Cite this article
Singh, P., Kim, YJ., Farh, M.EA. et al. Chryseobacterium panacis sp. nov., isolated from ginseng soil. Antonie van Leeuwenhoek 109, 187–196 (2016). https://doi.org/10.1007/s10482-015-0620-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-015-0620-2