Abstract
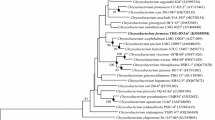
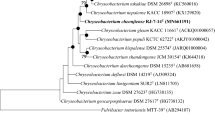
A Gram-stain negative, rod-shaped and non-endospore forming bacterium, designated strain YG4-6T, was isolated from Polytrichastrum formosum collected from Gawalong glacier in Tibet, China and characterized by using a polyphasic taxonomic approach. The predominant fatty acids of strain YG4-6T were identified as iso-C15:0 (29.3 %), summed feature 3 (C16:1 ω7c and/or C16:1 ω6c as defined by MIDI, 23.5 %) and iso-C17:0 3-OH (16.5 %). The major polar lipids were found to consist of five unidentified aminolipids and three unidentified lipids. Strain YG4-6T was found to contain MK-6 as the dominant menaquinone and the G+C content of its genomic DNA was determined to be 37.3 mol%. The phylogenetic analysis based on 16S rRNA gene sequences showed that strain YG4-6T is affiliated to Chryseobacterium species, and its closest related species were Chryseobacterium aahli T68T (97.9 % sequence similarity), Chryseobacterium zeae JM-1085T (97.8 % sequence similarity), Chryseobacterium yeoncheonense DCY67T (97.6 % sequence similarity) and Chryseobacterium soldanellicola NBRC 100864T (97.2 % sequence similarity). However, the DNA–DNA relatedness values between these strains and strain YG4-6T were found to be clearly below 70 %. Based on the phylogenetic inference and phenotypic data, strain YG4-6T is considered to represent a novel species of the genus Chryseobacterium, for which the name Chryseobacterium polytrichastri sp. nov. is proposed. The type strain is YG4-6T (=CGMCC 1.12491T = DSM 26899T). An emended description of the genus Chryseobacterium is also proposed.

Similar content being viewed by others
References
Behrendt U, Ulrich A, Schumann P (2008) Chryseobacterium gregarium sp. nov., isolated from decaying plant material. Int J Syst Evol Microbiol 58:1069–1074
Bernardet JF, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Bernardet JF, Vancanneyt M, Matte-Tailliez O, Grisez L, Tailliez P, Bizet C et al (2005) Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals. Syst Appl Microbiol 28:640–660
Breznak JA, Costilow RN (2007) Physicochemical factors in growth. In: Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 309–329
De Ley J (1970) Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol 101:738–754
Dong XZ, Cai MY (2001) Determination of biochemical properties. Manual for systematic identification of general bacteria. Science Press, Beijing, pp 370–398 (in Chinese)
Gillis M, Deley J, Decleene M (1970) Determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem 12:143–153
Ilardi P, Fernández J, Avendaňo-Herrera R (2009) Chryseobacterium piscicola sp. nov., isolated from diseased salmonid fish. Int J Syst Evol Microbiol 59:3001–3005
Im WT, Yang JF, Kim SY, Yi TH (2011) Chryseobacterium ginsenosidimutans sp. nov., a bacterium with ginsenoside-converting activity isolated from soil of a rhus vernicifera-cultivated field. Int J Syst Evol Microbiol 61:1430–1435
Kämpfer P, Vaneechoutte M, Lodders N, Baere TD, Avesani V, Janssens M et al (2009) Description of Chryseobacterium anthropi sp. nov. to accommodate clinical isolates biochemically similar to Kaistella koreensis and Chryseobacterium haifense, proposal to reclassify Kaistella koreensis asChryseobacterium koreense comb. nov. and emended description of the genus Chryseobacterium. Int J Syst Evol Microbiol 59:2421–2428
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H et al (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kämpfer P, Arun AB, Young CC, Chen WM, Sridhar KR, Rekha PD (2010) Chryseobacterium arthrosphaerae sp. nov., isolated from the faeces of the pill millipede Arthrosphaera magna Attems. Int J Syst Evol Microbiol 60:1765–1769
Kämpfer P, McInroy JA, Glaeser SP (2014) Chryseobacterium zeae sp. nov., Chryseobacterium arachidis sp. nov., and Chryseobacterium geocarposphaerae sp. nov., isolated from the rhizosphere environment. Antonie Van Leeuwenhoek 105:491–500
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Li YH, Liu QF, Liu Y, Zhu JN, Zhang Q (2011) Endophytic bacterial diversity in roots of Typha angustifolia L. in the constructed Beijing Cuihu Wetland (China). Res Microbiol 162:124–131
Loch TP, Faisal M (2014) Chryseobacterium aahli sp. nov., isolated from lake trout (Salvelinus namaycush) and brown trout (Salmo trutta), and emended descriptions of Chryseobacterium ginsenosidimutans and Chryseobacterium gregarium. Int J Syst Evol Microbiol 64:1573–1579
Marmur J, Doty P (1962) Determination of base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Marmur J, Schildkraut CL, Doty P (1961) The reversible denaturation of DNA and its use in studies of nucleic acid homologies and the biological relatedness of microorganisms. J Chim Phys 58:945–955
Montero-Calasanz M, Göker M, Rohde M, Spröer C, Schumann P, Busse HJ et al (2013) Chryseobacterium hispalense sp. nov., a plant growth-promoting bacterium isolated from a rainwater pond in an olive plant nursery and emendation of the species Chryseobacterium defluvii, Chryseobacterium indologenes, Chryseobacterium wanjuense and Chryseobacterium gregarium. Int J Syst Evol Microbiol 63:4386–4395
Park MS, Jung SR, Lee KH, Lee MS, Do JO, Kim SB et al (2006) Chryseobacterium soldanellicola sp. nov. and Chryseobacterium taeanense sp. nov., isolated from roots of sand-dune plants. Int J Syst Evol Microbiol 56:433–438
Park YJ, Son HM, Lee EH, Kim JH, Mavlonov GT, Choi KJ et al (2013) Chryseobacterium gwangjuense sp nov., isolated from soil. Int J Syst Evol Microbiol 63:4580–4585
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Ruijssenaars HJ, Hartsmans S (2001) Plate screening methods for the detection of polysaccharase-producing microorganisms. Appl Microbiol Biotechnol 55:143–149
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. DE, MIDI Inc., Newark
Shen FT, Kämpfer P, Young CC, Lai WA, Arun AB (2005) Chryseobacterium taichungense sp. nov., isolated from contaminated soil. Int J Syst Evol Microbiol 55:1301–1304
Smibert RM, Kreg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kämpfer P, Maiden MCJ et al (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tindall BJ (1990a) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130
Tindall BJ (1990b) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Letts 66:199–202
Tindall BJ, Sikorski J, Smibert RM, Kreig NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, 3rd edn. ASM Press, Washington, DC, pp 330–393
Van-An H, Kim YJ, Ngoc Lan N, Yang DC (2013) Chryseobacterium yeoncheonense sp nov., with ginsenoside converting activity isolated from soil of a ginseng field. Arch Microbiol 195:463–471
Vandamme P, Bernardet JF, Segers P, Kersters K, Holmes B (1994) New perspectives in the classification of the Flavobacteria—description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom rev. Int J Syst Bacteriol 44:827–831
Vaneechoutte M, Kämpfer P, De Baere T, Avesani V, Janssens M, Wauters G (2007) Chryseobacterium hominis sp. nov., to accommodate clinical isolates biochemically similar to CDC groups II-h and II-c. Int J Syst Evol Microbiol 57:2623–2628
Wu YF, Wu QL, Liu SJ (2013) Chryseobacterium taihuense sp nov., isolated from a eutrophic lake, and emended descriptions of the genus Chryseobacterium, Chryseobacterium taiwanense, Chryseobacterium jejuense and Chryseobacterium indoltheticum. Int J Syst Evol Microbiol 63:913–919
Yoon JH, Kang SJ, Oh TK (2007) Chryseobacterium daeguense sp nov., isolated from wastewater of a textile dye works. Int J Syst Evol Microbiol 57:1355–1359
Acknowledgment
This work was funded by the Scientific Research Program of National Natural Science Foundation of China (No. 31100004 and No. 31470136).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10482_2014_338_MOESM1_ESM.docx
The phylogenetic trees made by neighbour-joining and maximum parsimony methods, transmission electron micrographs and the TLC of the total polar lipids of strain YG4-6T are available as supplementary figures with the online version of this paper. (DOCX 1697 kb)
Rights and permissions
About this article
Cite this article
Chen, X.Y., Zhao, R., Chen, Z.L. et al. Chryseobacterium polytrichastri sp. nov., isolated from a moss (Polytrichastrum formosum), and emended description of the genus Chryseobacterium . Antonie van Leeuwenhoek 107, 403–410 (2015). https://doi.org/10.1007/s10482-014-0338-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0338-6