Abstract
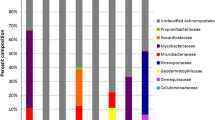
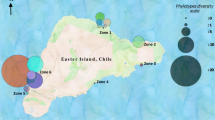
The Gulf of California is a coastal marine ecosystem characterized as having abundant biological resources and a high level of endemism. In this work we report the isolation and characterization of Actinobacteria from different sites in the western Gulf of California. We collected 126 sediment samples and isolated on average 3.1–38.3 Actinobacterial strains from each sample. Phylogenetic analysis of 136 strains identified them as members of the genera Actinomadura, Micromonospora, Nocardiopsis, Nonomuraea, Saccharomonospora, Salinispora, Streptomyces and Verrucosispora. These strains were grouped into 26–56 operational taxonomic units (OTUs) based on 16S rRNA gene sequence identities of 98–100 %. At 98 % sequence identity, three OTUs appear to represent new taxa while nine (35 %) have only been reported from marine environments. Sixty-three strains required seawater for growth. These fell into two OTUs at the 98 % identity level and include one that failed to produce aerial hyphae and was only distantly related (≤95.5 % 16S identity) to any previously cultured Streptomyces sp. Phylogenetic analyses of ketosynthase domains associated with polyketide synthase genes revealed sequences that ranged from 55 to 99 % nucleotide identity to experimentally characterized biosynthetic pathways suggesting that some may be associated with the production of new secondary metabolites. These results indicate that marine sediments from the Gulf of California harbor diverse Actinobacterial taxa with the potential to produce new secondary metabolites.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aparicio JF, Molnár I, Schwecke T, König A, Haydock SF, Khaw LE, Staunton J, Leadlay PF (1996) Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16
Bevitt DJ, Cortes J, Haydock SF, Leadlay PF (1992) 6-Deoxyerythronolide-B synthase-2 from Saccharopolyspora erythraea. Cloning of the structural gene, sequence analysis and inferred domain structure of the multifuncional enzyme. Eur J Biochem 204:39–49
Bull AT, Stach JEM (2007) Marine Actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499
Carreño AL, Helenes J (2002) Geology and ages of the islands. In: Case TJ, Cody ML, Equarra E (eds) A new Island biogeography of the sea of cortés. Oxford University Press, Oxford, pp 14–40
Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH (2004) Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod 67:1356–1367
Donadio S, Katz L (1992) Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51–60
Edlund A, Loesgen S, Fenical W, Jensen PR (2011) Geographic distribution of secondary metabolite genes in the marine actinomycete Salinispora arenicola. Appl Environ Microbiol 77:5916–5925
Fenical W, Jensen PR (2006) Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2:666–673
Freel KC, Nam SJ, Fenical W, Jensen PR (2011) Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol 77:7261–7270
Freel KC, Edlund A, Jensen PR (2012) Microdiversity and evidence for high dispersal rates in the marine actinomycete ‘Salinispora pacifica’. Environ Microbiol 14:480–493
Ginolhac A, Jarrin C, Robe P, PerriËre G, Vogel T, Simonet P, Nalin R (2005) Type I polyketide synthases may have evolved through horizontal gene transfer. J Mol Evol 60:716–725
Gontang EA, Fenical W, Jensen PR (2007) Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol 73:3272–3282
Gontang EA, Gaudêncio SP, Fenical W, Jensen PR (2010) Sequence-based analysis of secondary-metabolite biosynthesis in marine Actinobacteria. Appl Environ Microbiol 76:2487–2499
Hall TA, Brown JW (2001) The ribonuclease P family. Methods Enzymol 341:56–77
Han SK, Nedashkovskaya OI, Mikhailov VV, Kim SB, Bae KS (2003) Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol 53:2061–2066
Helmke E, Weyland H (1984) Rhodococcusmarinonascens sp. nov., an actinomycete from the sea. Int J Syst Bacteriol 34:127–138
Hozzein WN, Goodfellow M (2007) Streptomyces synnematoformans sp. nov., a novel actinomycete isolated from a sand dune soil in Egypt. Int J Syst Evol Microbiol 57:2009–2013
Jensen PR, Mafnas C (2006) Biogeography of the marine actinomycete Salinispora. Environ Microbiol 8:1881–1888
Jensen PR, Williams PG, Oh D-C, Zeiger L, Fenical W (2007) Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol 73:1146–1152
Liu Z, Li Y, Zheng LQ, Huang YJ, Li WJ (2010) Saccharomonospora marina sp. nov., isolated from an ocean sediment of the East China Sea. Int J Syst Evol Microbiol 60:1854–1857
Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M (2005) Salinispora arenicola gen. nov., sp nov and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol 55:1759–1766
Maldonado LA, Fragoso-Yañez D, Pérez-García A, Rosellón-Druker J, Quintana ET (2009) Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 95:111–120
Mincer T, Jensen PR, Christopher A, Kauffman Fenical W (2002) Widespread and persistent populations of a major new marine Actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011
Nagata H, Ochiai K, Aotani Y, Ando K, Yoshida M, Takahashi I, Tamaoki T (1997) Lymphostin (LK6-A), a novel immunosuppressant from Streptomyces sp. KY11783: taxonomy of the producing organism, fermentation, isolation and biological activities. J Antibiot 50:537–542
Olano C, Méndez C, Salas JA (2009) Antitumor compounds from marine actinomycetes. Mar Drugs 7:210–248
Penn K, Jenkis C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Morre BS, Jensen PR (2009) Genomic islands link secondary metabolism to functional adaptation in marine Actinobacterias. ISMEJ 3:1193–1203
Powers EM (1995) Efficacy of the Ryu non-staining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl Environ Microbiol 61:3756–3758
Roden GI, Groves GW (1959) Recent oceanographic investigations in the Gulf of California. J Mar Res 18:10–35
Rossello-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67
Solano G, Rojas-Jimenez K, Jaspars M, Tamayo-Castillo G (2009) Study of the diversity of culturable actinomycetes in the North Pacific and Caribbean coasts of Costa Rica. Antonie Van Leeuwenhoek 96:71–78
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tian XP, Tang SK, Dong JD, Zhang YQ, Xu LH, Zhang S, Li WJ (2009a) Marinactinospora thermotolerans gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int J Syst Evol Microbiol 59:948–952
Tian XP, Zhi XY, Qiu YQ, Zhang YQ, Tang SK, Xu LH, Zhang S, Li WJ (2009b) Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China sea. Int J Syst Evol Microbiol 59:222–228
Walsh CT, O’Connor SE, Schneider TL (2003) Polyketide-nonribosomal peptide epothilone antitumor agents: the EpoA, B, C subunits. Ind Microbiol Biotechnol 30:448–455
Yi H, Schumann P, Sohn K, Chun J (2004) Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with l-ornithine and l-serine in the peptidoglycan. Int J Syst Evol Microbiol 54:1585–1589
Acknowledgments
We acknowledge financial support from the Universidad Autónoma de Baja California Internal Assembly (grants No. 357 and 369), the Consejo Nacional de Ciencia y Tecnología (México) for the pre-doctorate fellowship given to A.B.E (number 5892). P.J. acknowledges financial support from the National Institutes of Health (grant GM0886261) and the NOAA California Sea Grant College Program Project R/NMP-100 (Grant NA100AR4170060) through NOAA’S National Sea Grant College Program, U.S. Dept. of Commerce. We thank W. Fenical for facilitating the field collections, M. Woolery for LC/MS assistance and D. Guillen, H. Ocampo-Alvarez, C. Barrila, S. Gomez, N. Millán and M. Torres for help with strain preservation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Becerril-Espinosa, A., Freel, K.C., Jensen, P.R. et al. Marine Actinobacteria from the Gulf of California: diversity, abundance and secondary metabolite biosynthetic potential. Antonie van Leeuwenhoek 103, 809–819 (2013). https://doi.org/10.1007/s10482-012-9863-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9863-3