Abstract
While the concept that physical forces such as tension and compression are involved in mature tissue modeling is widely accepted, the role of these specific types of mechanical loading in the differentiation and maturation of uncommitted cell types like human mesenchymal stem cells (hMSCs) is currently unknown. We observed that hMSCs have the fundamental ability to distinguish between dynamic tensile and compressive loading by regulating distinct gene expression patterns and that these differences in gene expression can be related to conformational changes in cell shape and volume. Dynamic tension was found to regulate both fibroblastic and osteogenic associated genes while dynamic compression up-regulated genes associated with chondrogenesis. Identifying genes involved in the mechanotransduction of different modes of physical loading in hMSC may greatly enhance the ability to rationally design tissue regeneration systems to restore proper tissue function.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human bone marrow contains a population of multipotent cells known as mesenchymal stem cells (hMSCs) capable of replication as undifferentiated cells2 or differentiation into multiple cell types including osteoblasts, fibroblasts, and chondrocytes.1,2,5,18,32,41 These positive attributes makes them an attractive cell source for tissue engineering applications where multiple cell and tissue types are required to restore diseased or damaged organ function.2
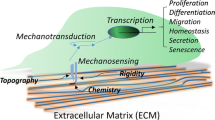
While hMSCs represent a novel form of cellular sourcing with many beneficial traits, the environmental conditions needed to develop functional tissue equivalents must be determined before employing stem cells in tissue engineering applications. There is evidence in all musculoskeletal tissues that mechanical loading affects the synthesis and the organization of extracellular matrix.3,17,33,34,45 Since the quality and intensity of local cellular loading in vivo is spatially and temporally complex, it is plausible that cells need appropriate mechanical cues in addition to biological signals from growth factors, cytokines, and extracellular matrix, to attain and sustain their mature biological characteristics. This, in conjunction with the capacity of mesenchymal cells to being driven down specific lineages, suggests that mechanical loading may play a critically important role in determining hMSC fate.
Historical data suggests that cells can distinguish between different forms of mechanical exposure. A particularly interesting distinction lies between octahedral normal stress (hydrostatic pressure) and octahedral shear stress (distortion): octahedral normal stress is a volume-changing stress, while octahedral shear is a shape-changing stress.8 In general, these two types of stress combine to form the total stress at a particular location. The utility of these classifications is supported by loading experiments on tissues where chondrogenic protein expression in cartilage is up-regulated in response to pressure,15,22 while structural protein expression is enhanced in fibrous connective tissues in response to distortion.4,22,23 While the concept that physical factors such as pressure and distortion are involved in tissue modeling and remodeling has become widely accepted, the majority of this evidence has been collected from differentiated cell types or tissue explants. Whether hMSCs can be manipulated down specific developmental lineages by load remains to be determined.
The primary objective of this study was to test the hypothesis that hMSCs have the fundamental ability to distinguish between different types of mechanical signals as evidenced by distinct gene expression patterns, and that these differences in gene expression can be related to specific components of mechanical stimulation: changes in cell shape and volume. To test this hypothesis, we utilized confocal microscopy to define cellular strains in response to discrete types of loading and oligonucleotide microarrays to identify genes that were differentially regulated by hMSCs after 24 h of either continuous dynamic pressure or distortional loading. Identifying the genes involved in the mechanotransduction of different modes of loading in hMSC may contribute to the success of physical or pharmacological therapies which utilize hMSC and greatly enhance the ability to rationally design tissue regeneration systems to restore proper tissue function.
Methods
Fabrication and Culture of Cell Seeded Constructs
Three-dimensional culture systems were employed to apply dynamic loading to hMSCs. Human bone marrow cells (Cambrex, Walkersville, MD) were expanded to third passage in tissue culture using media tested for support of stem cell proliferation (Poietics, BioWhittaker, Walkersville, MD).
For dynamic pressure, cells were suspended in 2% Improved Kelmar potassium alginate (ISP, San Diego, CA) in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, St. Louis, MO) at a cell density of 1 × 106 cells/mL. The alginate was then placed in dialysis tubing (Spectrum Lab, St. Rancho Dominguez, CA) and polymerized for 40 min in a HEPES buffered 102 mM CaCl2 solution. Cylindrical samples (6 mm OD × 2 mm height) were made using a 6 mm OD dermal biopsy punch (Miltex, York, PA) in combination with a custom cutting shim to ensure parallel faces.
For dynamic distortion, a bulk polymerizing CaCO3–GDL (d-glucono-δ-lactone) alginate system31 was utilized. A 2% Protanal LF200S alginate solution (FMC Biopolymer, Drammen, Norway) in Ca2+ and Mg2+ free Hank’s BSS provided the base of the copolymer and calcium carbonate was used as the cross-linking agent. The cross-linking density was maintained using a basic calcium ion to carboxyl molar ratio of 0.18 and a CaCO3 to GDL (Sigma, Eugene, OR) molar ratio of 0.5 was maintained to achieve a neutral pH value and similar mechanical properties as compressive samples.38 All solutions were heated to 37 °C and cells were suspended for a final cell density of 1 × 106 cells/mL. Tensile samples were cast (10 mm OD × 10 mm gauge length) utilizing a custom casting chamber to embed polyethylene rods (70 um pore size; 5 mm height) into both ends of the gel construct. Each polyethylene end-cap is fitted with an alloy set screw to allow tensile gripping of the cell-gel constructs in the bioreactor with a magnetic gripping system. Polymerization occurred for 10 min at room temperature before specimens were removed from the casting chamber.
After casting, both tensile and compressive cell constructs were rinsed 3 × 5 min in DMEM and cultured without load in a serum-free, defined media shown to maintain, but not promote, both fibroblastic and chondrogenic phenotypes38 for 24 h prior to dynamic loading. Cells were expanded, cultured, and loaded on separate occasions to produce biological replicates (n = 4) for each type of loading.
Dynamic Loading of Cell Seeded Constructs
Sample loading occurred in a custom bioreactor constructed from a high precision actuator (0.1 nm resolution) and motion controller assembly (PI Polytech, Auburn, MA), in series with a 1 kg load cell (Sensotec, Columbus, OH) housed at 37 °C in a humid 5% CO2–95% air incubator (Fig. 1). A set of custom loading chambers was employed to simultaneously load 28 samples in uniaxial unconfined compression (Fig. 1a) or 26 samples in uniaxial tension (Fig. 1b). Labview software (National Instruments, Austin, TX) was employed to control actuator position and to collect and analyze time dependant load and displacement data. All samples were placed in the base of the loading chamber and the top of the chamber was lowered at 1 μm increments via custom Labview® software until a 2 g preload ensured contact. Fresh serum-free media was added to chambers. Samples were then loaded in either uniaxial unconfined compression or tension under displacement controlled sinusoidal dynamic loading at 0.1 Hz frequency. The magnitude of the sine wave corresponded to ±5% bulk strain with a 5% offset (cycles between 0 and 10%) in compression or ±2.5% bulk strain with a 2.5% offset (cycles between 0 and 5%) in tension. Paired samples for each loading condition were placed in contact without load to act as negative controls.
Bioreactor for in vitro loading of cell-seeded constructs. Dynamic loading apparatus comprised of load cell (L), actuator (A), and loading chamber (C). Contact without load control chamber housed within testing incubator at left rear. Dynamic compression samples (n = 28) mounted on compression bioreactor base (a). Dynamic tensile loading specimen affixed to tension bioreactor base (b)
Cell Viability
To determine whether mechanical loading induced cell death, cell viability was compared after 24 h between loaded and unloaded samples. For each condition, a central 2 mm longitudinal section of each construct (n = 3 per condition) was placed in media containing 25 μg/mL calcein AM and 20 μg/mL ethidium homodimer (Molecular Probes, Eugene, OR) for 20 min at 37 °C and rinsed four times with PBS. Epi-fluorescence microscopy was utilized to visualize cells that fluoresce green (live) and red (dead). ImageJ software v1.3342 was utilized for automated cell counting.
Cell measurements were also performed to determine permanent changes in cell shape induced by dynamic loading. Measurements included cell area, perimeter, major and minor axis, cell alignment, and cell circularity as defined by the ratio of the minor to major axis (ranges from 0 = flat line to 1 = perfect circle). Statistical analysis of all samples was performed using student’s t-test analysis on JMP v5 (SAS, Cary, NC); p < 0.05 was considered to represent significance.
Confocal Microscopy
To determine the changes in cell volume and shape (deviatoric strain) during loading, confocal microscopy was performed on statically loaded samples from each condition. While under load, samples were incubated in 10% buffered formalin for 1 h, removed from loading chamber, embedded (Tissue-Tek® O.C.T. Compound, (Baxter, McGraw Park, IL) and snap frozen in liquid nitrogen. Cryosections (60 um) were cut, incubated for 5 min in PBS, and labeled at 37 °C for 10 min with 1 μg/mL CM-DiI (Molecular Probes, Eugene, OR), a fluorescent dye that labels cellular membrane, and washed in PBS for 5 min. Slides were coverslipped with a 1:1 dilution of Slowfade Antifade (component B) containing the nuclear counterstain DAPI (Molecular Probes) in PBS. Cells were visualized with a Zeiss LSM 510 confocal microscope at 20× magnification using a Plan-Apochromat objective. Multitrack configuration allowed dual tracking of DI positive staining (790 nm titanium/sapphire laser/Rhodamine filter) and DAPI (HeNe laser 543 nm/DAPI filter). Full thickness z-stack images were obtained at 1.0 μm intervals and averaging of 4 lines. Skeletal length, which corresponds to the maximum length of the object in three-dimensions, skeletal diameter, which is the maximum length perpendicular to the skeletal length, and volume measurements were performed (n = 131) on 3-D rendered z-stacks using Volocity 2.0 software (Improvision, Lexington, MA). Statistical analysis was performed using ANOVA with Tukey’s post-hoc analysis on JMP v5 (SAS, Cary, NC); p < 0.05 was considered to represent significance.
Gene Expression Analysis
To determine genes uniquely regulated by either pressure or distortion, each dynamically loaded sample was compared to its contact (0% strain) counterpart (n = 4 biological replicates per condition). Cells were released from alginate constructs by dissolving alginate in a solution of 55 mM Sodium citrate, 0.15 M HEPES, 25 mM NaCl (Sigma, Eugene, OR) for 10 min.31 Gene expression analysis consisted of total RNA isolation with DNase treatment (RNeasy® Mini kit, Qiagen, Valencia, CA), followed by amplification of 1 μg of total RNA (Message Amp II aRNA Kit, Ambion, Austin, TX). Fluorescently tagged cDNA was created by reverse transcription of 20 μg of total amplified RNA from each condition using amino-allyl dUTP (Sigma, Eugene, OR) and subsequent coupling to monofunctional NHS-ester Cy3 and Cy5 dyes (Amersham, Buckinghamshire, England). Each loading condition and its corresponding unloaded control were hybridized on human oligonucleotide microarrays (Gladstone Genomics, UCSF, CA) containing sequences for 21,329 human genes from the OPERON human genome, as previously described.28 Mean pixel intensity for each gene was quantified using a GenePix 4000B microarray scanner (Axon Instruments, Union City, CA) and GenePixPro software 6.0 (Axon Instruments, Union City, CA). Photomultiplier tube gains were balanced such that both channels had the same relative intensity per photon detected (count ratio = 1.0). Background fluorescence was subtracted at each wavelength and data was normalized by Lowess normalization28 using Acuity v4.0 software (Axon Instruments, Union City, CA) to correct for dye properties and labeling efficiencies.
Statistically significant changes in gene expression were determined utilizing Statistical Analysis of Microarrays (SAM) software (Stanford, Palo Alto, CA).48 Genes significantly up or down regulated by either dynamic pressure or dynamic distortion were determined by comparing the loaded gene expression levels to their contact without load controls using SAM software analysis for one class response. To determine statistically significant changes in gene expression between the normalized pressure and distortion loading regimes, SAM analysis for two-class, unpaired data was used. Pearson’s Hierarchical clustering was performed using Acuity v4.0 software (Axon Instruments, Union City, CA) to determine the correlation coefficient between genes regulated by dynamic pressure and distortion loading. Significantly different genes were linked to biological pathway data using GenMapp 2.0 beta (Gladstone Institute, University of California San Francisco, SF, CA)11 and gene ontology determined using MappFinder (Gladstone Institute, UCSF, SF, CA).13 Statistically significant changes in gene expression by a factor greater than 1.5 were considered of biological interest.
Quantitative RT-PCR
To confirm microarray gene expression patterns, quantitative RT-PCR was performed on a select number of genes. Synthetic oligonucleotide primers and probes designed to specifically amplify and detect Aggrecan, collagen type I alpha I, versican, ADAM-TS4 and GAPDH were purchased (Assays-on-Demand, Applied Biosystems). Primers and probe for BMP-6 were designed using PrimerExpress-1.7 software (Applied Biosystems, Foster City, CA). cDNA was made from 1 ng of amplified mRNA (Message Amp II aRNA, Ambion, Austin, TX) using TaqMan Reverse Transcription reagent kits (Applied Biosystems) and with ThermoScript Reverse Transcription System (Invitrogen, Carlsbad, CA). Real-time quantitative RT-PCR was done on an ABI 7700 Sequence Detector using TaqMan reagents (Applied Biosystems) following recommended protocols. Reverse transcription reactions were performed multiple times and analyzed by real-time quantitative PCR in triplicate. Results were normalized to GAPDH levels and fold change calculated using the nonpaired 2−ΔΔCt method.37 Statistical analysis was performed using ANOVA with Tukey’s post-hoc analysis on JMP v5 (SAS, Cary, NC); p ≤ 0.05 was considered to represent significance.
Results
Dynamic Loading Regimes did not Alter Cell Viability or Total Cell Numbers
To enable proper comparison between the various samples, we first determined whether loading conditions affected the overall cell number or viability. We found that cell viability was over 80% in all conditions (82.44 ± 4.56%). We found no significant changes in cell number or viability after 24 h of either dynamic pressure (p = 0.229) or dynamic distortion (p = 0.461).
Cellular Deformations During Compressive and Tensile Loading
We observed that during compressive loading cycles, cells were compacted by the surrounding alginate hydrogel. When compared to its contact control, the increased pressure during compressive loading resulted in a decrease in cell skeletal length (0.711; p < 0.0001), skeletal diameter (0.838; p = 0.003), volume (0.688; p < 0.0001), and surface area (0.660; p < 0.0001) (Fig. 2). Cell strain was calculated from changes in cell diameter in the axis of loading and revealed that a 10% bulk strain of the alginate produced a 9.02 ± 0.66% strain on the cellular level. The volume decreases observed in hMSCs compressed in alginate were similar to those found in compression studies of intact cartilage.46
Tension and compression devices delivered different types of mechanical forces at the cellular level. Quantification of 3D rendered confocal images of human mesenchymal stem cells in alginate under either compressive or tensile bulk deformations. All measurements normalized to unloaded control. Mean ± SEM (n = 131) (left). Representative 3D rendered confocal images (right)
Conversely, we noted that during the tensile loading cycles, cells were stretched by the surrounding alginate matrix. When compared to the contact control (Fig. 2), tensile loading produced an increase in cell skeletal length (1.638; p < 0.0001), volume (1.99; p < 0.0001), and surface area (1.417; p < 0.0001), and a decrease in skeletal diameter (0.641; p = 0.003). Tensile loading also produced a significant increase in cell distortion as measured by the ratio of the skeletal length to the cell diameter (p < 0.0001) when compared to the compressed cells or contact only controls.
Dynamic Loading Increases Cell Size Post-load
After both compressive and tensile loading, we observed a significant increase in cell size compared to unloaded controls. This increase in size was assessed by cell area (pressure: p = 0.0009; distortion: p < 0.0001), perimeter (pressure: p = 0.0007; distortion: p < 0.0001), major axis (pressure: p = 0.0004; distortion: p < 0.0001), and minor axis (pressure: p = 0.0019; distortion: p < 0.0001). While cell size increased, there was no change in cell circularity (pressure: p = 0.133; distortion: p = 0.886) of the cells after dynamic loading was removed.
Dynamic Compression Regulates Genes Associated with Chondrocyte Proliferation and Chondrogenesis
When comparing cDNA from dynamically compressed cells to unloaded control cells, 20,617 genes were detected for statistical analysis. Comparison between paired sets (n = 4) of oligonucleotide microarrays by SAM software revealed that 861 genes were statistically regulated by compression (204 genes were up-regulated and 657 were down-regulated) (80% correlation; median number of false positives = 0.342; delta = 4.47; Table 1).
Gene ontology analysis was performed to identify general cellular processes likely to be affected by dynamic compression of hMSCs. The genes up-regulated by dynamic pressure were associated with cellular processes including the regulation of actin cytoskeleton and organogenesis while down-regulated genes were associated with features of the Wnt signaling pathway, integrin mediated cell adhesion, and apoptosis.
Many of the significantly regulated genes have previously been reported as promoting chondrogenesis. These genes included the up-regulation of alpha-catenin, cathepsin B, aggrecan (p ≤ 0.0001), BMP-6 (p = 0.001), ADAM-TS4 (p = 0.001), and the significant down-regulation in FGF-1, IL-1β, collagen 1α1 (p = 0.0002) and versican (p = 0.016). Selected chondrogenic genes were confirmed using quantitative RT-PCR (n = 6) and there was no significant difference between RT-PCR and microarray gene expression levels for all genes assayed (p > 0.561).
Dynamic Tension Up-regulated Genes Associated with Bone Formation and Inhibited Chondrogenesis
When comparing cDNA from dynamically tensioned cells to unloaded control cells, 18,773 genes were detected for statistical analysis. Comparison between paired sets (n = 4) of oligonucleotide microarrays by SAM software revealed that 784 genes were statistically regulated by tension (365 genes were up-regulated and 419 were down-regulated) (80% correlation; median number of false positives = 2.662; delta = 0.505; Table 2). Dynamic tension resulted in a significant increase in ossification-associated genes including the calcitonin receptor, beta-catenin, ADAM-TS, Runx2, and Wnt-8. Dynamic tension also produced a significant decrease in chondrogenesis associated genes BMPR1a, and the Wnt receptor Frizzled-7. Gene ontology analysis revealed that genes up-regulated by dynamic distortion were associated with G-protein signaling while down-regulated genes were associated with the TGF-β and insulin signaling pathways.
Dynamic Pressure and Distortion Induce Distinct Gene Expression Patterns and Signaling Events
When comparing dynamic pressure and tension directly, 15,274 genes were adequately detectable for statistical analysis. Comparison between normalized oligonucleotide microarrays by SAM software revealed that 638 genes were statistically regulated (263 genes were up-regulated and 375 were down-regulated) (80% correlation; median number of false positives = 0.495; delta = 1.754; Table 3). Pearson’s centered clustering was performed on normalized array data (n = 4/condition) to produce a 2 branched dendrogram separating dynamic pressure and distortion arrays by a correlation coefficient of 0.372.
Dynamic tension significantly up-regulated osteogenic genes including bone morphogenetic protein 1 (BMP1), alkaline phosphatase, and Nell1, and genes encoding for known regulators of cell volume including Aquaporin 3 (AQP3). Dynamic pressure showed significant up-regulation of chondrogenic-associated genes Wnt3, Wnt5a, hyaluronan synthase 2 (HAS2), and Collagen type XI, alpha 2 (COL11A2). Some other genes of interest related to chondrogenesis were not changed in a statistically significant manner (e.g., Col II, fold change 0.9; and Sox 9, fold change 8.0).
Gene ontology analysis revealed 15 significant ontologies (p < 0.05) between pressure and distortion including G-protein coupled receptor signaling, protein biosynthesis and signal transduction.
Discussion
The objective of our study was to determine whether human mesenchymal stem cells are capable of responding to physical stimuli, and specifically, to determine whether tensile and compressive forces elicit a differential response. During dynamic tension, we observed large increases in cell volume, with high levels of cellular distortion caused by cyclic octahedral shear stresses that correlated to decreases in chondrogenic gene expression. With dynamic compression we noted relatively smaller decreases in cell volume, low levels of cellular distortion, and an up-regulation of chondrogenesis associated genes. These results are consistent with prior reports that cyclic motion stimulates intramembranous ossification in regions of low to moderate tensile strain and hydrostatic tensile stress, while chondrogenesis is stimulated in areas of hydrostatic compressive stress.6
In particular, we observed that dynamic pressure stimulates and that dynamic distortion antagonizes Wnt mediated chondrogenesis. Wnts are secreted glycoproteins that activate signal transduction pathways to control a wide variety of cellular processes. Two categories of Wnts have been described, canonical Wnts such as Wnt-1, -3a, and -8 that stabilize beta-catenin via their receptor frizzled, and non-canonical Wnts such as Wnt-4, -5a, and -11 that activate the Wnt/calcium signaling pathway via activation of protein kinase C.9,14,16,26,50 The influence of Wnt signaling on chondrogenic differentiation varies with the specific Wnt and the cell stage.19 For example, Wnt-4 and Wnt-8 inhibit chondroprogenitor differentiation but also enhance transition of proliferative chondrocytes to the prehypertrophic stage,14,49 while this transition is inhibited by overexpression of Wnt-3a, Wnt-5a, and frizzled-7 receptor.20,21 We observed that dynamic tension upregulated Wnt-8 and down regulated frizzled-7, suggesting this stimulation may facilitate the transition to prehypertrophy and endochondral ossification. Conversely, when compared to tension, Wnt-3a and Wnt-5a were upregulated by pressure thereby supporting the chondrocyte proliferation and chondrogenesis.
Trends in alpha- and beta-catenin also implicate differing roles for compression and tension in supporting either cartilage or bone formation. These proteins influence intercellular adhesions by complexing with the cytosolic component of the linker protein cadherin. They also play a role in modulating the canonical Wnt signaling pathway. Alpha-catenin gene expression was upregulated by pressure. This protein has a role in chondrocyte differentiation by disrupting the transcriptional activity of beta-catenin.12 For example, genetic inactivation of β-catenin in MSCs in vitro leads to chondrocyte differentiation, even under culture conditions that would promote bone.12 Conversely, β-catenin gene expression was upregulated by dynamic tension, which serves to stabilize cell-to-cell adhesions and thereby inhibit chondrogenesis.24,35
An up-regulation of cathepsin B secondary to dynamic pressure supports chondrocyte proliferation. Cathepsin B is a lysosomal cysteine proteinase that can cleave aggrecan at a site near that for MMP-3 (stromelysin-1). It is also active against fibronectin and can activate pro-stromelysin in vitro.39 Cathepsin B is scarcely produced in differentiated chondrocytes but is excessively produced by de-differentiated chondrocytes in vitro or in osteoarthritis where it is characteristic of chondrocyte cloning.52 Up-regulation of cathepsin B by itself is not sufficient for enhanced cell secretion into the extracellular matrix.39 Rather, the stimulus for cell secretion appears to be adjacent matrix depletion. Since chondrocytes need to degrade matrix to provide space for proliferation, it has been hypothesized that cathepsin B plays a physiologic role in the normal cell division cycle.
Down-regulation of FGF and IL-1 during dynamic pressure also support chondrocyte proliferation. FGF and its receptor FGFR act as negative regulators of chondrocyte growth.27,43 IL-1a is also a suppresser of chondrocyte proliferation and survival,44 and both inhibits proteoglycan synthesis and up-regulates mediators of matrix damage.
In contrast to pressure, distortion promoted gene expression associated with bone formation. Genes upregulated by distortion included ADAMTS, calcitonin receptor, and Nell1. ADAMTSs are aggrecanases that are cell secreted and incorporated into the extracellular matrix, and are thought to play a role in inflammation by degrading extracellular matrix.30 Since proteoglycan removal is necessary for bone formation, ADAMTS are thought to be important for mineralization and have been observed to be an osteogenic marker that accumulates in matrix around osteoblasts during differentiation.36 The calcitonin receptor is an osteoclast differentiation marker.29 Osteoclast differentiation is thought to be initiated before the deposition of cartilage or bone matrix as calcitonin-binding cells appear before cartilage and bone formation in the area of ectopic bone formation induced by BMP.25 Nell1 is a protein strongly present in cells of neural crest origin residing within the craniofacial complex.10 It has been observed to be upregulated in abnormally fusing cranial sutures and is specific for osteochondral progenitor cells. Nell1 protein stimulates bone formation similarly to BMP-7, and has been reported to promote accelerated differentiation of osteoblastic lineage cells.51
We noted a number of novel genes to be mechanically induced by each loading regime. One potentially interesting gene family is the aquaporins. Aquaporins are membrane bound water channel proteins that function as selective pores. They have been identified in many tissues including human cartilage.40 Aquaporins (AQP) are categorized based on their selectivity: AQP0, AQP1, AQP2, AQP4, AQP5, AQP6, and AQP8 pass only water while AQP3, AQP7, and AQP9 have been shown to pass neutral solutes like glycerol and urea.40 The function of aquaporins in articular cartilage is largely unknown though it is speculated that a potential role of the AQP gene family is in the movement of ECM and metabolic water across the membranes of chondrocytes for the purpose of cell volume regulation and homeostasis.47 Our findings have interesting implications to the role of these channel proteins in cartilage function and homeostasis during loading.
AQP1 is expressed in human chondrocytes located in the deep zone of articular cartilage where fluid pressurization dominates7 while AQP3 is present in the deep zone and strongest at the tidemark.47 Cartilage is rigidly held at the bone–cartilage interface at the tidemark that is characterized by high interfacial shear strains and little to no compressive strains.7 The high expression of AQP3 in an area of high tensile load correlated well with our findings that AQP3 is significantly up-regulated by dynamic distortion and decreased by dynamic pressure. Other aquaporins—AQP2 and AQP5—showed a non-significant trend towards up-regulation during dynamic distortion and decrease during pressure.
In summary, the load-specific biological responses of hMSC suggest that these cells can distinguish between different mechanical environments and that they modulate their gene expression patterns accordingly. The changes in cell volume and distortion produced by the different loading regimes suggest that hMSCs may be guided along different cell differentiation pathways by controlling the mechanical forces experienced by the cell, and ultimately, form different tissue types based at least partly on their loading history (Fig. 3). By identifying the genes involved in these distinct downstream adaptations to mechanical load, we may greatly enhance the ability to rationally design tissue regeneration systems to restore proper tissue function.
References
Barry F., R. E. Boynton, B. Liu, et al. 2001 Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp. Cell. Res. 268(2), 189–200. doi:10.1006/excr.2001.5278
Bianco P., M. Riminucci, S. Gronthos, et al. 2001 Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells 19(3), 180–192. doi:10.1634/stemcells.19-3-180
Booth F. W., E. W. Gould 1975 Effects of training and disuse on connective tissue. Exerc. Sport Sci. Rev. 3, 83–112. doi:10.1249/00003677-197500030-00006
Breen E. C. 2000 Mechanical strain increases type I collagen expression in pulmonary fibroblasts in vitro. J. Appl. Physiol. 88(1), 203–209
Caplan A. I., M. Elyaderani, Y. Mochizuki, et al., 1997 Principles of cartilage repair and regeneration. Clin. Orthop. 342, 254–269. doi:10.1097/00003086-199709000-00033
Carter D. R., G. S. Beaupre, N. J. Giori, et al. 1998 Mechanobiology of skeletal regeneration. Clin. Orthop. Relat. Res. 331(355 Suppl), S41–S55. doi:10.1097/00003086-199810001-00006
Carter D. R., G. S. Beaupre, M. Wong, et al., 2004 The mechanobiology of articular cartilage development and degeneration. Clin. Orthop. Relat. Res. 427(Suppl), S69–S77. doi:10.1097/01.blo.0000144970.05107.7e
Carter D. R., G. S. Beaupre 2001 Skeletal Function and Form. New York: Cambridge University Press
Church V. L., P. Francis-West 2002 Wnt signalling during limb development. Int. J. Dev. Biol. 46(7), 927–936
Cowan C. M., S. Cheng, K. Ting, et al. 2006 Nell-1 induced bone formation within the distracted intermaxillary suture. Bone 38(1), 48–58. doi:10.1016/j.bone.2005.06.023
Dahlquist K. D., N. Salomonis, K. Vranizan, et al. 2002 GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat. Genet. 31(1), 19–20. doi:10.1038/ng0502-19
Day T. F., X. Guo, L. Garrett-Beal, et al. 2005 Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 8(5), 739–750. doi:10.1016/j.devcel.2005.03.016
Doniger S. W., N. Salomonis, K. D. Dahlquist, et al. 2003 MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 4(1), R7. doi:10.1186/gb-2003-4-1-r7
Enomoto-Iwamoto M., J. Kitagaki, E. Koyama, et al. 2002 The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev. Biol. 251(1), 142–156. doi:10.1006/dbio.2002.0802
Evanko S. P., K. G. Vogel 1993 Proteoglycan synthesis in fetal tendon is differentially regulated by cyclic compression in vitro. Arch. Biochem. Biophys. 307(1), 153–164. doi:10.1006/abbi.1993.1574
Fischer L., G. Boland, R. S. Tuan 2002 Wnt signaling during BMP-2 stimulation of mesenchymal chondrogenesis. J. Cell. Biochem. 84(4), 816–831. doi:10.1002/jcb.10091
Forwood M. R., C. H. Turner 1995 Skeletal adaptations to mechanical usage: results from tibial loading studies in rats. Bone 17(4 Suppl), 197S–205S. doi:10.1016/8756-3282(95)00292-L
Friedenstein A. J., K. V. Petrakova, A. I. Kurolesova, et al. 1968 Heterotopic of bone marrow. Analysis of precursor cells for osteogenic and hematopoietic tissues. Transplantation 6(2), 230–247. doi:10.1097/00007890-196803000-00009
Goldring M. B., K. Tsuchimochi, K. Ijiri 2006 The control of chondrogenesis. J. Cell. Biochem. 97(1), 33–44. doi:10.1002/jcb.20652
Hartmann C. 2002 Wnt-signaling and skeletogenesis. J. Musculoskelet. Neuronal Interact. 2(3), 274–276
Hartmann C., C. J. Tabin 2000 Dual roles of Wnt signaling during chondrogenesis in the chicken limb. Development 127(14), 3141–3159
Howard P. S., U. Kucich, R. Taliwal, et al. 1998 Mechanical forces alter extracellular matrix synthesis by human periodontal ligament fibroblasts. J. Periodontal Res. 33(8), 500–508
Hsieh A. H., R. L. Sah, K. L. Paul Sung 2002 Biomechanical regulation of type I collagen gene expression in ACLs in organ culture. J. Orthop. Res. 20(2), 325–331. doi:10.1016/S0736-0266(01)00112-7
Hwang S. G., S. S. Yu, J. H. Ryu, et al. 2005 Regulation of beta-catenin signaling and maintenance of chondrocyte differentiation by ubiquitin-independent proteasomal degradation of alpha-catenin. J. Biol. Chem. 280(13), 12758–12765. doi:10.1074/jbc.M413367200
Irie K., C. Alpaslan, K. Takahashi, et al. 2003 Osteoclast differentiation in ectopic bone formation induced by recombinant human bone morphogenetic protein 2 (rhBMP-2). J. Bone Miner. Metab. 21(6), 363–369. doi:10.1007/s00774-003-0430-x
Ishikawa Y. 2005 Wnt signaling and orthopedic diseases. Am. J. Pathol. 167(1), 1–3
Jang J. H., C. P. Chung 2005 Fibronectin-mediated adhesion rescues cell cycle arrest induced by fibroblast growth factor-1 by decreased expression of p21(cip/waf) in human chondrocytes. In Vitro Cell Dev. Biol. Anim. 41(5–6), 126–129. doi:10.1290/0412082.1
Kelly, D. L., and A. Rizzino. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol. Reprod. Dev. 56(2):113–123, 2000. doi:10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q
Kim M. S., C. J. Day, C. I. Selinger, et al. 2006 MCP-1-induced human osteoclast-like cells are tartrate-resistant acid phosphatase, NFATc1, and calcitonin receptor-positive but require receptor activator of NFkappaB ligand for bone resorption. J. Biol. Chem. 281(2), 1274–1285. doi:10.1074/jbc.M510156200
Kuno K., Y. Okada, H. Kawashima, et al. 2000 ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 478(3), 241–245. doi:10.1016/S0014-5793(00)01854-8
Kuo C. K., P. X. Ma 2001 Ionically crosslinked alginate hydrogels as scaffolds for tissue engineering: part 1. Structure, gelation rate and mechanical properties. Biomaterials 22(6), 511–521. doi:10.1016/S0142-9612(00)00201-5
Kuznetsov S. A., M. H. Mankani, S. Gronthos, et al. 2001 Circulating skeletal stem cells. J. Cell. Biol. 153(5), 1133–1140. doi:10.1083/jcb.153.5.1133
Lacroix D., P. J. Prendergast 2002 A mechano-regulation model for tissue differentiation during fracture healing: analysis of gap size and loading. J. Biomech. 35(9), 1163–1171. doi:10.1016/S0021-9290(02)00086-6
Lee D. A., D. L. Bader 1997 Compressive strains at physiological frequencies influence the metabolism of chondrocytes seeded in agarose. J. Orthop. Res. 15(2), 181–188. doi:10.1002/jor.1100150205
Lee H. S., S. J. Millward-Sadler, M. O. Wright, et al. 2000 Integrin and mechanosensitive ion channel-dependent tyrosine phosphorylation of focal adhesion proteins and beta-catenin in human articular chondrocytes after mechanical stimulation. J. Bone Miner. Res. 15(8), 1501–1509. doi:10.1359/jbmr.2000.15.8.1501
Lind T., N. McKie, M. Wendel, et al. 2005 The hyalectan degrading ADAMTS-1 enzyme is expressed by osteoblasts and up-regulated at regions of new bone formation. Bone 36(3), 408–417. doi:10.1016/j.bone.2004.11.008
Livak K. J., T. D. Schmittgen 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4), 402–408. doi:10.1006/meth.2001.1262
Lodie T. A., C. E. Blickarz, T. J. Devarakonda, et al. 2002 Systematic analysis of reportedly distinct populations of multipotent bone marrow-derived stem cells reveals a lack of distinction. Tissue Eng. 8(5), 739–751. doi:10.1089/10763270260424105
Mehraban F., M. H. Tindal, M. M. Proffitt, et al. 1997 Temporal pattern of cysteine endopeptidase (cathepsin B) expression in cartilage and synovium from rabbit knees with experimental osteoarthritis: gene expression in chondrocytes in response to interleukin-1 and matrix depletion. Ann. Rheum. Dis. 56(2), 108–115
Mobasheri A., E. Trujillo, S. Bell, et al. 2004 Aquaporin water channels AQP1 and AQP3, are expressed in equine articular chondrocytes. Vet. J. 168(2), 143–150. doi:10.1016/j.tvjl.2003.08.001
Pittenger M. F., A. M. Mackay, S. C. Beck, et al. 1999 Multilineage potential of adult human mesenchymal stem cells. Science 284(5411), 143–147. doi:10.1126/science.284.5411.143
Rasband, W. ImagJ: Image Processing and Analysis in JAVA. 2004. Available at http://rsb.info.nih.gov/ij/index.html.
Sahni M., D. C. Ambrosetti, A. Mansukhani, et al. 1999 FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 13(11), 1361–1366. doi:10.1101/gad.13.11.1361
Schuerwegh A. J., E. J. Dombrecht, W. J. Stevens, et al. 2003 Influence of pro-inflammatory (IL-1 alpha, IL-6, TNF-alpha, IFN-gamma) and anti-inflammatory (IL-4) cytokines on chondrocyte function. Osteoarthritis Cartilage 11(9), 681–687. doi:10.1016/S1063-4584(03)00156-0
Storey E., S. A. Feik 1982 Remodelling of bone and bones. Effects of altered mechanical stress on anlages. Br. J. Exp. Pathol. 63(2), 184–193
Szafranski J. D., A. J. Grodzinsky, E. Burger, et al. 2004 Chondrocyte mechanotransduction: effects of compression on deformation of intracellular organelles and relevance to cellular biosynthesis. Osteoarthritis Cartilage 12(12), 937–946. doi:10.1016/j.joca.2004.08.004
Trujillo E., T. Gonzalez, R. Marin, et al. 2004 Human articular chondrocytes, synoviocytes and synovial microvessels express aquaporin water channels; upregulation of AQP1 in rheumatoid arthritis. Histol. Histopathol. 19(2), 435–444
Tusher V. G., R. Tibshirani, G. Chu 2001 Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98(9), 5116–5121. doi:10.1073/pnas.091062498
Yano F., F. Kugimiya, S. Ohba, et al. 2005 The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9-dependent manner. Biochem. Biophys. Res. Commun. 333(4), 1300–1308. doi:10.1016/j.bbrc.2005.06.041
Yates K. E., S. Shortkroff, R. G. Reish 2005 Wnt influence on chondrocyte differentiation and cartilage function. DNA Cell Biol. 24(7), 446–457. doi:10.1089/dna.2005.24.446
Zhang X., S. Kuroda, D. Carpenter, et al. 2002 Craniosynostosis in transgenic mice overexpressing Nell-1. J. Clin. Invest. 110(6), 861–870
Zwicky R., A. Baici 2000 Cytoskeletal architecture and cathepsin B trafficking in human articular chondrocytes. Histochem. Cell. Biol. 114(5), 363–372
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Haudenschild, A.K., Hsieh, A.H., Kapila, S. et al. Pressure and Distortion Regulate Human Mesenchymal Stem Cell Gene Expression. Ann Biomed Eng 37, 492–502 (2009). https://doi.org/10.1007/s10439-008-9629-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-008-9629-2