Abstract
Free-living snakes may carry a broad range of pathogens that differ considerably by geographical locations and source species. The biological role of wildlife snakes for transmission of bacteria and helminths is hardly explored. The purpose of the study was a snapshot on the zoonotic agents load in free-living snakes found dead in a landscape park in central Poland. A total of 16 dead free-living snakes identified as European grass snake (Natrix natrix, N = 15) and a smooth snake (Coronella austriaca, N = 1) were tested. Abdominal organs were used for bacteriological testing, whereas, parasites were searched for in the remaining carcasses. Fourteen (87.5 %) individuals were positive for Salmonella spp., whereas, trematodes were found in 13 out of 16 samples (81.3 %). A total of 33 isolates representing 11 Salmonella serovars or antigenic forms were observed with Salmonella enterica subsp. diarizonae (IIIb) 38:r:z being the most frequent. The trematodes were recognized as Alaria alata mesocercariae (Distomum musculorum suis, DMS). Additionally, unidentified nematodes were detected in four samples. No Yersinia spp. and Mycobacterium spp. were found. The results indicate huge burden of Salmonella spp. and muscle parasites carriage by snakes present in the natural environment and indicate possible zoonotic and epidemiological impact. It justifies our concept of free-living snakes as possible source and vector of pathogens, especially for omnivores and scavengers foraging on snake corpses. Finding of rare and diverse Salmonella serovars and unidentified nematodes proves the need for in-depth studies and systematic approach to reveal the role of free-living reptiles in epidemiology of infectious agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epidemiology of infectious and parasitic diseases includes extensive transmission cycles with well-defined sources, vectors, and routes of spread of causative agents. Numerous research continuously provide new data for upgrading of state of the art. They usually focus on details in well-known areas, but less affordable or inconvenient elements of epidemiological puzzle remain unrecognized. To exemplify, Salmonella occurrence in reptiles and their role in pathogen transmission to humans are commonly known, but the vast majority of research efforts is focussed on Salmonella epidemiology in farm animals.
A few snakes occur in the natural environment of Poland. All species, including European grass snake (Natrix natrix) and smooth snake (Coronella austriaca) are under protection. That fact hampers research efforts unless the clinical threats occur (Mihalca et al. 2010; Sting et al. 2013). Definitely, wildlife snakes carry a broad range of bacteria and parasites that differs considerably by geographical locations and source species (Borkovcová and Kopřiva 2005; Fitzgerald et al. 2013; Hacioglu and Tosunoglu 2014; Lukac et al. 2015; Scheelings et al. 2011; Schmidt et al. 2014; Shimalov and Shimalov 2000). Salmonella spp., being considered natural component of reptile gut flora, is one of the most frequently noted (Goupil et al. 2012; Kuroki et al. 2013; Prapasarakul et al. 2012; Richards et al. 2004; Scheelings et al. 2011). Wildlife snakes are often reported as host of new parasites or record their new geographical location (Borkovcová and Kopřiva 2005; Halajian et al. 2013; Santoro et al. 2011; Shimalov and Shimalov 2000; Yildirimhan et al. 2007).
The biological role of free-living snakes for transmission of bacteria and helminths is hardly explored. One might presume them as paratenic hosts (Duscher et al. 2015; Halajian et al. 2013; Mohl et al. 2009), although public health consequences of direct contact with carrier reptile itself or its’ contaminated environment shall not be neglected (Mohl et al. 2009; Zając et al. 2013).
Since there are considerable gaps in evidence-based knowledge of European grass and smooth snakes in transmission of infectious and parasitic agents (Schmidt et al. 2014), the purpose of the study was a snapshot on the zoonotic agents load in free-living snakes found dead in a landscape park in central Poland.
Materials and methods
Sampling
Sixteen dead free-living snakes were collected in “Gostynińsko-Włocławski Park Krajobrazowy” (a landscape park in central Poland) in autumn 2014. The location of animal collection (15.8-km long asphalt forest road), injuries, and the state of the corpses indicated on the cause of death due to traffic accidents that happened within few (fresh blood) or several hours (dried body tissues). The corpses were submitted for laboratory testing on two occasions: 5th Oct. (10 animals) and 12th Oct. (6 animals). The animals were identified as European grass snake (Natrix natrix, N = 15) and a smooth snake (Coronella austriaca). Due to the severe damages of the bodies, full-scale necropsy was not feasible, and two types of samples were collected for laboratory diagnostics. Abdominal organs were used for bacteriological testing, whereas, parasites were searched for in the remaining carcasses.
Bacteriological testing
Fragments of internal organs were homogenized and incubated with oxalic acid. Following a cycle of saline flushes, the sample was streaked on Stonebrink and Petragnani media and cultured for Mycobacterium spp.
The remaining fragments of organs were pooled with the intestines and homogenized in buffered peptone water (BPW, 1:10 w/v ratio). One milliliter of the homogenate was inoculated in peptone sorbitol bile broth (PSB, 10 ml, incubation in 5 °C for 2 weeks) and further on cefsulodin-irgasan-novobiocin agar (CIN, 28 °C, 48 h) for Yersinia enterocolitica isolation.
The remaining volume of BWP homogenate was cultured for Salmonella spp. according to EN ISO 6579:2002/A1:2007. Besides EU official methodology, preincubated culture was streaked on chromogenic RAPID Salmonella agar (Bio-Rad). Up to five presumptive colonies from each selective medium plate were confirmed for Salmonella with PCR targeting invA and serotyped according to White-Kauffmann-Le Minor scheme (Grimont and Weill 2007). From each sample, a single isolate showing distinct serological properties was used for further analyses. These included Salmonella subspecies identification with multiplex PCR and antimicrobial susceptibility testing (minimal inhibitory concentration, Sensititre®, Trek D. S.) as previously described (Zając et al. 2013).
Parasitological testing
Muscle samples were mechanically extracted from unskinned snake’s carcasses, although in a few cases, the whole dried carcass was tested. The individual samples were chopped and digested using pepsin and hydrochloric acid by the magnetic stirrer method (MSM) followed by microscopy as required by reference Trichinella detection method (Commission Regulation (EC) No 2075/2005). The found parasites were preserved in 96 % ethanol for further species identification with PCR assay. Nematode DNA extract (DNA IQ System, Promega) was used to amplify two fragments of 18S ribosomal DNA (rDNA) gene as described by Blaxter et al. (1998). Both amplicons were sequenced, aligned (Geneious v. 7.1.0), and compared with the GenBank database.
Results
Of the 16 snakes tested, 14 (87.5 %) were positive for Salmonella spp., 13 (81.3 %) carried Distomum musculorum suis, and larvae of unrecognized helminths were noted in four cases (Table 1). No Yersinia spp. and Mycobacterium spp. were found.
A total of 33 isolates representing 11 Salmonella serovars or distinct antigenic forms were observed with Salmonella enterica subsp. diarizonae (IIIb) 38:r:z being the most frequent (9 N. natrix). Up to four different Salmonella serovars were found in a single animal. The serovars belonged to subspecies diarizonae (N = 9) and enterica (N = 2). Twenty-four out of 26 Salmonella showed microbiological resistance to streptomycin (MIC range from 32–128 mg/L), whereas, the isolates were susceptible to all tested β-lactams and cephalosporins, phenicols, quinolones and fluoroquinolones, aminoglycosides, tetracycline, sulphonamides, trimethoprim, and colistin.
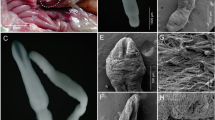
Thirteen snakes (81.3 %) were infected with mesocercaria of Alaria alata, usually named Distomum musculorum suis Duncker, 1896 (DMS). The number of trematodes ranged from three to 45 per analyzed sample (Table 1). Additionally, yet unidentified nematodes (up to four per sample, electron microscopy results not shown) were found in four carcasses, including three snakes with DMS co-infestation. The amplification and sequencing of 18S rDNA have not defined the nematode genus, and the definite identification is still under investigation.
Discussion
Both tested snake species are under strict protection according to international and national law. Grass snake is the most common snake found in Poland living in the environments close to the inland waters and preying mainly on amphibians. The smooth snake prefers dry and sunlight areas and feeds mainly on lizards but also amphibians and small rodents. Every year, numerous snakes are killed on road because of their migration and the need for basking. Availability of snake carcasses on roads provides an opportunity of easy meal for omnivores and scavengers, i.e., red foxes or wild boars. Some studies have reported over 50 % of snake carcasses removal from the roads within 8 hours and up to 1 hour from forest roads (DeGregorio et al. 2010). This fact indicates road-killed snakes as a potential source of bacterial infection and parasitic infestation for wild scavengers and omnivores.
Since deliberate capture of protected reptiles is prohibited, the coincidental acquisition of number of dead animals gave a huge opportunity to put some insight on carriage of selected bacteria and parasites in native population. Scientific literature (WoS search, August 2015) on the topic is sparse, and current results might be considered basic and unique.
The study showed high occurrence of diverse Salmonella serovars belonging mostly to Salmonella (S.) enterica subspecies diarizonae. This includes the second frequent atypical serological form (21:l,v:-) with missing second phase antigens. It is in line with German study reporting unknown and rare Salmonella serovars (Krautwald-Junghanns et al. 2013). The observed frequency of Salmonella carriage was higher than found in the literature (Krautwald-Junghanns et al. 2013; Kuroki et al. 2013) but comparable to the studies focused on captive animals (Geue and Loschner 2002; Prapasarakul et al. 2012). Our results clearly confirm free-living snakes as reservoir of numerous S. enterica subspecies and serovars (Grupka et al. 2006; Krautwald-Junghanns et al. 2013; Kuroki et al. 2013). The tangible proofs for that are finding of S. enterica subspecies diarizonae 40:i:z53,z54 (not listed in White-Kauffmann-Le Minor scheme), and S. enterica subspecies diarizonae 48:k:z57 already reported in free-living adders in Germany (Krautwald-Junghanns et al. 2013) and humans in Poland (Issenhuth-Jeanjean et al. 2014). The later striking example draws attention to possible epidemiological links and public health consequences of free-living snake-associated salmonellosis.
Although both analyzed snake species were obtained from the same location, Salmonella serovars noted in Coronella austriaca did not occur in Natrix natrix. The same serovars found in several grass snake individuals might suggest their direct contact, i.e., during mating or basking, as main route of Salmonella transmission. Research done on lizard population proved that bacteria are transmitted from host to host around the social network, rather than adjacent individuals from some common source (Bull et al. 2012). The observed differences also can result from different habitats and prey preferences of both snakes species. Influence of food intake on Salmonella prevalence is hard to estimate, but some studies indicate on this route as a way of transmission of new serovars for an individual (Fuller et al. 2008).
Antimicrobial resistance testing of bacteria isolated from free-living snakes is seldom reported. Further, reptile-associated Salmonella show less resistance compared to the ones from farm animals (Zając et al. 2013). The observed streptomycin resistance is usually the sole resistance being reported in often pan-susceptible Salmonella of snake origin (Kuroki et al. 2013; Schmidt et al. 2014; Smith et al. 2012; Zając et al. 2013).
Despite non-tuberculosis mycobacteria and Yersina spp. ware not found, there are studies reporting those pathogens in reptiles (Mitchell 2012; Soldati et al. 2004).
The study demonstrated high prevalence of DMS, the trematode also found in game animals in Poland (Chmurzyńska et al. 2013). The obtained data suggest that free-living snakes may act as vectors of parasites for wild scavengers and omnivores, i.e., wild boars. High numbers of DMS reported in free-living snakes (Mohl et al. 2009; Shimalov and Shimalov 2000) might be related with their prey on amphibians. Noteworthy, both the prevalence and enumeration of parasites might have been reduced by several factors. These include sample condition (limited volume, dried or decayed carcasses) and methodology. The digestion method was arbitrary chosen based on the assumption that small rodents, a component of snake’s diet, are reservoirs of Trichinella (Flis 2011). Unexpected Alaria alata mesocercariae might have been efficiently detected with migration method, since a percentage of DMS is destroyed by acid concentration and pepsine or stuck on the sieve (Riehn et al. 2010). These, along with the finding of still unidentified nematodes, show the area for further investigation and possible knowledge gaps.
In conclusion, current results indicate huge burden of Salmonella spp. and muscle parasites carriage by snakes present in the natural environment. Since clinical consequences might be neglected, the findings draw attention to possible zoonotic and epidemiological impacts. Snake carcasses removal from roads by omnivorous, i.e., wild boars, may lead to high numbers of this animals infected with Alaria alata and Salmonella spp. The prevalence of Salmonella in wild boars in Poland reached 6.0 % (unpublished study), and some infrequent S. enterica serovars were noted (i.e., S. Tripoli). It might justify our concept of free-living snakes as possible source and vector of both pathogens. Finding of rare Salmonella serovars, still unidentified nematodes prove the need for further studies and systematic approach to reveal the role of reptiles in epidemiology of infectious agents.
References
Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM, Vida JT, Thomas K (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392:71–75. doi:10.1038/32160
Borkovcová M, Kopřiva J (2005) Parasitic helminths of reptiles (Reptilia) in South Moravia (Czech Republic). Parasitol Res 95:77–78. doi:10.1007/s00436-004-1258-6
Bull CM, Godfrey SS, Gordon DM (2012) Social networks and the spread of Salmonella in a sleepy lizard population. Mol Ecol 21:4386–4392. doi:10.1111/j.1365-294X.2012.05653.x
Chmurzyńska E, Różycki M, Bilska-Zając E, Karamon J, Cencek T (2013) Alaria alata jako potencjalne zagrożenie zdrowia ludzi—występowanie i rozpoznawanie. Życie Wet 88:780–784
DeGregorio BA, Nordberg EJ, Stepanoff KE, Hill JE (2010) Patterns of snake road mortality on an isolated barrier island. Herpetol Cons Biol 5:441–448
Duscher GG, Leschnik M, Fuehrer H-P, Joachim A (2015) Wildlife reservoirs for vector-borne canine, feline and zoonotic infections in Austria. Int J Parasitol 4:88–96. doi:10.1016/j.ijppaw.2014.12.001
Fitzgerald KT, Shipley BK, Newquist KL, Vera R, Flood AA (2013) Additional observations and notes on the natural history of the prairie rattlesnake (Crotalus viridis) in Colorado. Top Companion Anim M 28:167–176. doi:10.1053/j.tcam.2013.09.008
Flis M (2011) Występowanie włośnicy u dzików i świń w Polsce w latach 2003–2009. Medycyna Wet 67:470–473
Fuller CC, Jawahir SL, Leano FT, Bidol SA, Signs K, Davis C, Holmes Y, Morgan J, Teltow G, Jones B, Sexton RB, Davis GL, Braden CR, Patel NJ, Deasy MP 3rd, Smith KE (2008) A multi-state Salmonella Typhimurium outbreak associated with frozen vacuum-packed rodents used to feed snakes. Zoonoses Public Health 55:481–487. doi:10.1111/j.1863-2378.2008.01118.x
Geue L, Loschner U (2002) Salmonella enterica in reptiles of German and Austrian origin. Vet Microbiol 84:79–91. doi:10.1016/S0378-1135(01)00437-0
Goupil BA, Trent AM, Bender J, Olsen KE, Morningstar BR, Wunschmann A (2012) A longitudinal study of Salmonella from snakes used in a public outreach program. J Zoo Wildl Med 43:836–841. doi:10.1638/2011-0281R1.1
Grimont PAD, Weill F-X (2007) Antigenic formulae of Salmonella serovars, 9th edn. WHO Collaborating Centre for Research on Salmonella, Institute Pasteur, Paris
Grupka LM, Ramsay EC, Bemis DA (2006) Salmonella surveillance in a collection of rattlesnakes (Crotalus spp.). J Zoo Wildl Med 37:306–312. doi:10.1638/05-059.1
Hacioglu N, Tosunoglu M (2014) Determination of antimicrobial and heavy metal resistance profiles of some bacteria isolated from aquatic amphibian and reptile species. Environ Monit Assess 186:407–413. doi:10.1007/s10661-013-3385-y
Halajian A, Bursey CR, Goldberg SR, Gol SMA (2013) Helminth parasites of the European glass lizard, Pseudopus apodus (Squamata: Anguidae), and European grass snake, Natrix natrix (Serpentes: Colubridae), from Iran. Comp Parasitol 80:151–156. doi:10.1654/4588.1
Issenhuth-Jeanjean S, Roggentin P, Mikoleit M, Guibourdenche M, de Pinna E, Nair S, Fields PI, Weill F-X (2014) Supplement 2008–2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res Microbiol 165:526–530. doi:10.1016/j.resmic.2014.07.004
Krautwald-Junghanns ME, Stenkat J, Szabo I, Ortlieb F, Blindow I, Neul AK, Pees M, Schmidt V (2013) Characterization of Salmonella isolated from captive and free-living snakes in Germany. Berl Munch Tierarztl Wochenschr 126:209–215
Kuroki T, Ishihara T, Furukawa I, Okatani AT, Kato Y (2013) Prevalence of Salmonella in wild snakes in Japan. Jpn J Infect Dis 66:295–298. doi:10.7883/yoken.66.295
Lukac M, Pedersen K, Prukner-Radovcic E (2015) Prevalence of Salmonella in captive reptiles from Croatia. J Zoo Wildl Med 46:234–240. doi:10.1638/2014-0098R1.1
Mihalca AD, Miclaus V, Lefkaditis M (2010) Pulmonary Lesions caused by the nematode Rhabdias fuscovenosa in a Grass Snake, Natrix natrix. J Wildl Dis 46:678–681. doi:10.7589/0090-3558-46.2.678
Mitchell MA (2012) Mycobacterial infections in reptiles. Vet Clin North Am Exot Anim Pract 15(101–111):vii. doi:10.1016/j.cvex.2011.10.002
Mohl K, Grosse K, Hamedy A, Wuste T, Kabelitz P, Lucker E (2009) Biology of Alaria spp. and human exposition risk to Alaria mesocercariae—a review. Parasitol Res 105:1–15. doi:10.1007/s00436-009-1444-7
Prapasarakul N, Pulsrikarn C, Vasaruchapong T, Lekcharoen P, Chanchaithong P, Lugsomya K, Keschumras N, Thanomsuksinchai N, Tanchiangsai K, Tummaruk P (2012) Salmonella serovar distribution in cobras (Naja kaouthia), snake-food species, and farm workers at Queen Saovabha Snake Park, Thailand. J Vet Diagn Invest 24:288–294. doi:10.1177/1040638711434110
Richards JM, Brown JD, Kelly TR, Fountain AL, Sleeman JM (2004) Absence of detectable Salmonella cloacal shedding in free-living reptiles on admission to the wildlife center of Virginia. J Zoo Wildlife Med 35:562–563. doi:10.1638/03-070
Riehn K, Hamedy A, Grosse K, Zeitler L, Lücker E (2010) A novel detection method for Alaria alata mesocercariae in meat. Parasit Res 107:213–220. doi:10.1007/s00436-010-1853-7
Santoro M, Tkach VV, Mattiucci S, Kinsella JM, Nascetti G (2011) Renifer aniarum (Digenea: Reniferidae), an introduced North American parasite in grass snakes Natrix natrix in Calabria, southern Italy. Dis Aquat Org 95:233–240. doi:10.3354/dao02365
Scheelings TF, Lightfoot D, Holz P (2011) Prevalence of Salmonella in Australian reptiles. J Wildl Dis 47:1–11. doi:10.7589/0090-3558-47.1.1
Schmidt V, Mock R, Burgkhardt E, Junghanns A, Ortlieb F, Szabo I, Marschang R, Blindow I, Krautwald-Junghanns ME (2014) Cloacal aerobic bacterial flora and absence of viruses in free-living slow worms (Anguis fragilis), grass snakes (Natrix natrix) and European Adders (Vipera berus) from Germany. Ecohealth 11:571–580. doi:10.1007/s10393-014-0947-6
Shimalov VV, Shimalov VT (2000) Helminth fauna of snakes (Reptilia, Serpentes) in Belorussian Polesye. Parasitol Res 86:340–341
Smith KF, Yabsley MJ, Sanchez S, Casey CL, Behrens MD, Hernandez SM (2012) Salmonella isolates from wild-caught Tokay Geckos (Gekko gecko) imported to the U.S. from Indonesia. Vector Borne Zoonotic Dis 12:575–582. doi:10.1089/vbz.2011.0899
Soldati G, Lu ZH, Vaughan L, Polkinghorne A, Zimmermann DR, Huder JB, Pospischil A (2004) Detection of Mycobacteria and Chlamydiae in granulomatous inflammation of reptiles: a retrospective study. Vet Pathol Online 41:388–397. doi:10.1354/vp.41-4-388
Sting R, Ackermann D, Blazey B, Rabsch W, Szabo I (2013) Salmonella infections in reptiles-prevalence, serovar spectrum and impact on animal health. Berl Munch Tierarztl Wochenschr 126:202–208
Yildirimhan HS, Bursey CR, Goldberg SR (2007) Helminth parasites of the grass snake, Natrix natrix, and the dice snake, Natrix tessellata (Serpentes: Colubridae), from Turkey. Comp Parasitol 74:343–354. doi:10.1654/4285.1
Zając M, Wasyl D, Hoszowski A, Le Hello S, Szulowski K (2013) Genetic lineages of Salmonella enterica serovar Kentucky spreading in pet reptiles. Vet Microbiol 166:686–689. doi:10.1016/j.vetmic.2013.07.023
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zając, M., Wasyl, D., Różycki, M. et al. Free-living snakes as a source and possible vector of Salmonella spp. and parasites. Eur J Wildl Res 62, 161–166 (2016). https://doi.org/10.1007/s10344-016-0988-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10344-016-0988-y