Abstract
Loss of genetic variation from genetic drift during population bottlenecks has been shown for many species. Red deer (Cervus elaphus) may have been exposed to bottlenecks due to founder events during postglacial colonisation in the early Holocene and during known population reductions in the eighteenth and nineteenth centuries. In this study, we assess loss of genetic variation in Scandinavian red deer due to potential bottlenecks by comparing microsatellite (n = 14) and mitochondrial DNA variation in the Norwegian and Swedish populations with the Scottish, Lithuanian and Hungarian populations. Bottlenecks are also assessed from the M ratio of populations, heterozygosity excess and from hierarchical Bayesian analyses of their demographic history. Strong genetic drift and differentiation was identified in both Scandinavian populations. Microsatellite variation was lower in both Scandinavian populations compared with the other European populations and mitochondrial DNA variation was especially low in the Swedish population where only one unique haplotype was observed. Loss of microsatellite alleles was demonstrated by low M ratios in all populations except the Hungarian. M ratios’ were especially low in the Scandinavian populations, indicating additional or more severe bottlenecks. Heterozygosity excess compared with the expectation from the number of observed microsatellite alleles suggested a recent bottleneck of low severity in the Norwegian population. Hierarchical Bayesian coalescent analyses consistently yielded estimates of a large ancestral and a small current population size in all investigated European populations and suggested the onset of population decline to be between 5,000 and 10,000 years ago, which coincide well with postglacial colonisation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Loss of genetic variation from genetic drift during population bottlenecks has been demonstrated for many species (Weber et al. 2000; Vila et al. 2003; Goossens et al. 2006), as a result of processes such as founder events and isolation after postglacial colonisation (Hewitt 2000; Hewitt 2004) or as a consequence of human activities during the last centuries (Vila et al. 2003; Weber et al. 2004; Nystrom et al. 2006). However, even though rare alleles are soon lost during a bottleneck, heterozygosity is only reduced when it is severe or long-lasting (Maruyama and Fuerst 1985; Allendorf 1986). Few empirical studies of wild populations have thus provided convincing evidence of significant loss of heterozygosity through bottlenecks (Amos and Harwood 1998; Amos and Balmford 2001).
During the Pleistocene ice ages, cold climate and expansion of the polar ice sheets compressed vegetation zones and species ranges into southern refuges towards the Equator (Andersen and Borns 1994; Hewitt 1996, 2000). With the interglacial periods’ shifts to warmer climate and retreat of permafrost and continental ice caps, species expanded northwards along a leading edge, involving founder events and loss of genetic variation in the northern areas (Hewitt 1996, 2000, 2004). In Europe, red deer (Cervus elaphus) first appeared in the Pleistocene during the Cromerian interglacial approximately half a million years ago and fossil records suggest a subsequent distribution changing with the glacial cycles (Flerov 1952; Kurtèn 1968; Lister 1984; Lister 1993). During the last glacial maximum (LGM) around 22–18,000 years before present (BP), most of northern Europe was covered by a thick ice sheet (Andersen and Borns 1994; Clark and Mix 2002; Clark et al. 2009). Red deer phylogeography suggests two genetically distinct LGM refuges; (1) the Iberian Peninsula and possibly the Italian Peninsula and (2) the Balkans (Skog et al. 2009). LGM-dated archaeological findings coincide with these but suggest wider refuges which include areas in southwestern France and the Carpathians (Sommer and Nadachowski 2006; Sommer et al. 2008).
As the ice sheet retreated through several oscillations until around 8,500 BP (Andersen and Borns 1994), on each side of the Alps red deer expanded from (1) the Iberian refuge into western and northern Europe and (2) from the Balkan refuge into eastern Europe (Sommer et al. 2008; Skog et al. 2009). The glacial reduction of the sea level had created land bridges that in northern Europe connected the continent with the British Isles and the southern part of the Scandinavian Peninsula (Andersen and Borns 1994). After invasion via these land bridges, red deer appeared around 9,500 BP in Britain (Lister 1984) and southern Sweden (Jonsson 1995; Liljegren and Ekström 1996) and around 6,700 BP in western Norway (Rosvold 2006). Around 8,000 BP, a sea level rise from melting ice flooded the land bridges and the southern part of Scandinavia and the British Isles became isolated from the continent (Andersen and Borns 1994; Jonsson 1995). Archaeological findings indicate wide prehistoric distributions in Britain (9,500–5,000 BP, Lister 1984) and Sweden (9,000–3,800 BP, Ahlèn 1965) while in Norway the vast majority of prehistoric findings (6,700–2,000 BP) have been made along the west coast (Lønnberg 1906; Ahlèn 1965; Rosvold 2006; Hufthammer 2006).
Today, the European red deer is divided into two distinct genetic lineages, one western and one eastern (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). Populations located today in past LGM refuges have higher levels of genetic variation than populations within areas of postglacial colonisation (Skog et al. 2009). In addition, genetic differentiation has been estimated on many different geographical scales by allozymes (Hartl et al. 1990; Strandgaard and Simonsen 1993; Herzog and Gehle 2001), mtDNA (Hartl et al. 2005) and microsatellite markers (Zachos et al. 2003; Feulner et al. 2004; Kuehn et al. 2004). In these studies, genetic differentiation was related to geography or isolation by distance (Hartl et al. 1990; Herzog and Gehle 2001; Kuehn et al. 2004) or to effects of anthropogenic influences like habitat fragmentation (Kuehn et al. 2003; Hartl et al. 2005), selective hunting or translocations between different populations (Hartl et al. 1991; Hartl et al. 2003).
During the Middle Ages, the Norwegian red deer population had a wide distribution, as indicated by records of comprehensive export of skin and antlers and frequent finds of red deer remains in the waste deposits of many large cities (Collett 1909; Grieg 1909; Collett 1912). In more recent times, there are written records of an abundant population across southern Norway in the sixteenth and seventeenth centuries (Claussøn Friis 1599; Pontoppidan 1753). During the eighteenth century, the Norwegian population was severely reduced and in the nineteenth century it was limited to a few localities along the west coast, counting only a few hundred individuals at the most extreme (Collett 1909; Ingebrigtsen 1924). In Sweden, written records indicate a wide red deer distribution before the eighteenth century, but extirpation from most localities occurred during the eighteenth and nineteenth centuries (Ahlèn 1965). Afterwards, indigenous Swedish red deer were only found in one southernmost area (Scania), fluctuating between 50 and 100 individuals until recently (Lønnberg 1906; Ahlèn 1965). Captive bred red deer of indigenous, presumed indigenous and unknown non-indigenous origin have since 1950 been introduced into localities outside Scandia (Ahlèn 1965; Gyllensten et al. 1983; Whalstöm 1996). The indigenous Swedish population have remained relatively isolated (Gill 1990). The introduced localities are, however, not particularly differentiated genetically from the Scandia population (Gyllensten et al. 1983). Separate red deer of unknown, non-indigenous origin have in addition, since 1950, been suspected of escaping from captivity in areas at far distances from Scandia (Whalstöm 1996). The Scandia population increased to around 200 individuals in the 1950s and around 400 in 1989 (Lavsund 1975; Lavsund 1990), and today counts 1,200–1,500 individuals before the yearly hunt (personal communication, Anders Jarnemo, Swedish University of Agricultural Sciences). The Norwegian population has expanded much more drastically both demographically and spatially during the last century, and particularly the last three decades (Gill 1990). Only one translocation is known, into a hereto isolated island population (Haanes et al. 2010). Today, red deer are common in most parts of southern and central Norway and the population counts from 100,000–120,000 individuals (Langvatn 1988; Forchhammer et al. 1998; Langvatn 1998).
Previous studies indicate a generally low level of microsatellite variation in Norwegian red deer (Røed 1998) and that allozyme variation is absent and low in the Norwegian and Swedish populations, respectively (Gyllensten et al. 1983). In this study, we compare the level of genetic variation at 14 microsatellite markers and one mtDNA sequence in the Scandinavian red deer populations with the levels in other European populations to address whether genetic variation has been lost due to bottlenecks since postglacial colonisation. In addition, we use specific tests for bottlenecks and assess the demographic history of each population through estimates of ancient and present population sizes and time since onset of any change.
Methods and materials
Sampling and laboratory procedures
From 2001 to 2005, muscle tissue and blood samples were obtained from Norwegian (n = 20, three regions), Swedish (n = 19, two localities), Scottish (n = 20, 12 localities), Lithuanian (n = 20, two regions) and Hungarian (n = 20, three regions) red deer (Fig. 1). The three non-Scandinavian populations were chosen for comparison since they are situated in areas of comparable postglacial colonisation. The Scottish and Hungarian populations each lie within the areas of postglacial colonisation from the Iberian and Balkan glacial refuge, respectively (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). The Scottish population is an island population that is comparable to the Scandinavian ones in terms of postglacial flooding of the LGM land bridges to continental Europe. The Lithuanian population, by comparison, has been landlocked since colonisation and the phylogeographic pattern implies that it too was colonised from the Iberian glacial refuge. The Swedish samples are from the isolated indigenous sanctuary (n = 11) in central Scandia (Ahlèn 1965; Gyllensten et al. 1983; Gill 1990) and from one locality at Hunneberg (n = 8) where red deer have been introduced from this sanctuary. The areas sampled in Norway all lie within the area of distribution recorded during the beginning of last century (Collett 1909; Ingebrigtsen 1924) and today represent three of five subpopulations (overall F st = 0.08, Haanes et al. 2010). The Scottish samples were collected from 12 estates along the Central-West Highlands (overall F st = 0.02, Pérez-Espona et al. 2008). The Hungarian samples were collected from three well-separated areas.
Genomic DNA was isolated from muscle tissue (Qiagen, DNeasy KIT) and genotyping performed using the following 14 dinucleotide microsatellite loci: CSSM03 (Moore et al. 1994); OarCP26 (Ede et al. 1995); RT5 (Wilson et al. 1997); SRCRSP10 (Bhebhe et al. 1994); NVHRT73 and NVHRT48 (Røed and Midthjell 1998); McM58 (Hulme et al. 1994); OarFCB193 and OarFCB304 (Buchanan and Crawford 1993) and BM5004, BM888, BMC1009, BM4208 and BM4107 (Bishop et al. 1994). Loci were amplified on a GeneAmp PCR System 9600 (Applied Biosystems, CA, USA) in 10 μL reaction mixtures with 30–60 ng of genomic template DNA, 2 pmol of each primer, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl, 0.2 mM dNTP and 0.5 U of AmpliTaq DNA polymerase (Applied Biosystems). An initial denaturation at 94°C for 5 min was followed by 30 cycles of amplification with 1 min at 95°C, 30 s at 55°C and 1 min at 72°C and a final 10 min extension at 72°C. PCR products were then separated by size using capillary electrophoresis (ABI PRISM 310) and electromorphs genotyped with GENOTYPER1.1.1 (both Applied Biosystems).
A 463 base-pair region of the mitochondrial D-loop adjacent to the tRNApro gene was amplified using the primers L15394 and H15947 (Flagstad and Røed 2003). Amplifications were performed in 10 l volumes containing 1.5 mM MgCl2, 200 μM of each dNTP, 4 pmol of each primer and 0.5 U of AmpliTaq DNA polymerase (Applied Biosystems). Thirty-five cycles of amplification with 30 s at 94 C, 30 s at 60 C and 45 s at 72 C were preceded by a 2-min pre-denaturation step and followed by a final 7-min extension. Sequencing was performed using BigDye terminator cycle sequencing chemistry on an ABI 3100 instrument, and sequences aligned manually using SeqScape version 2.0 (Applied Biosystems).
Statistical analyses
Genetic diversity and differentiation
Deviations from Hardy–Weinberg equilibrium were assessed for each population through exact tests across the 14 microsatellite loci using GENEPOP 3.4 with default settings (Rousset and Raymond 1995). Significance levels were sequentially Bonferroni adjusted for repeated tests (Rice 1989). To address the level of genetic variation and investigate for differences among populations in the microsatellite loci, we used FSTAT 2.9.3 (Goudet 2001) to calculate allelic richness (El Mousadik and Petit 1996) and gene diversity (Nei 1987) for each population across loci. Comparisons were also made with the allelic richness and gene diversity recorded in other studied populations. In four loci (OarCP26, OarFCB304, BM888 and BM4208), genotypes were obtained from Dr. Frank Zachos for nine populations in Serbia and Romania (n ∈ [6, 26], from Feulner et al. 2004), which lie within the area suggested as the Balkan glacial refuge (Sommer et al. 2008; Skog et al. 2009). In three loci, genotypes were obtained from Dr. Javier Peréz-González for one Iberian population (OarCP26, n = 27; and OarFCB304, OarFCB193, n = 240) in San Pedro (from Peréz-González et al. 2010), which lie within the area suggested as the Iberian glacial refuge (Sommer et al. 2008; Skog et al. 2009).
Genetic variation in the mtDNA control region was assessed from haplotype number (nh) and diversity (h), nucleotide diversity (nd) and Wattersons’ theta (θ), calculated using ARLEQUIN 3.0 (Excoffier et al. 2005). Neutrality tests (Tajima’s D) were also performed in ARLEQUIN. In addition, the distribution of haplotypes and relationships among haplotypes were visualised through a statistical parsimony network analysis using TCS 1.13 (Clement et al. 2000). Observed haplotype sequences (463 bp) were blasted in GeneBank (99% similarity criterion). To get a network as coherent as possible, eight European red deer sequences with known origins within Europe were downloaded and included: AF291886 and AF291887, AF291888, AF291889 (Italian, Norwegian and Spanish, respectively; Randi et al. 2001) and DQ386107–DQ386110 (all Scottish; Nussey et al. 2006).
Genetic structure among populations was addressed through F statistics (Weir 1996), calculated using FSTAT for the microsatellite data and through an analysis of molecular variance (Excoffier et al. 1992) in ARLEQUIN 3.0 for the mtDNA data, excluding indels and estimating significance from 3,000 permutations. F statistics were also used to address inbreeding (F is) and the overall F st within each separate population (theta) and the subsequent F is (f) when theta is accounted for. Genetic structure was further assessed through individual-based factorial correspondence analysis (FCA) as implemented in GENETIX 4.05.2 (Belkhir et al. 2004) and through Bayesian assessment without any prior information on sampling localities, as implemented in STRUCTURE 2.3 (Pritchard et al. 2000). In STRUCTURE, for each of a different number of genetic clusters (K ∈ [1, 7]), an admixture model (α = 1, α max = 50), with correlated allele frequencies (Falush et al. 2003), 100,000 burn-in cycles and 500,000 Monte Carlo Markov Chains (MCMC) iterations was run 10 times. Average posterior probability among runs and standard error were calculated for each K value. The main genetic structure was interpreted from ΔK, which is negatively related to the often increasing variance among runs and higher posterior probabilities of higher K values and which can be used to identify major breakpoints (Evanno et al. 2005).
Bottlenecks and demographic histories
Bottlenecks of short duration or low severity may involve loss of alleles without any significant reduction of heterozygosity, and are followed by a transient period of allele deficiency compared with what is expected from the observed heterozygosity (Nei et al. 1975; Maruyama and Fuerst 1985). The probability of a recent bottleneck was addressed in each population by testing if the observed heterozygosity (unbiased gene diversity) was higher than expected from the observed numbers of alleles across microsatellite loci, using 10,000 simulations to estimate the expected heterozygosity and a one-tailed Wilcoxon test implemented in the software BOTTLENECK (Cornuet and Luikart 1996). Since most microsatellites fit a two-phase model of mutation (TPM) with 80–95% of the stepwise mutation model (SMM; Kimura and Ohta 1978) better than a strict one-step model (Di Rienzo et al. 1994), we used TPM models with 80% and 90% SMM.
Furthermore, the M ratio (Garza and Williamson 2001) was calculated for each locus as the ratio between the number of alleles observed in each separate population compared to the total number of alleles observed across all the sampled populations and averaged across loci. Only the gaps within the repeat size range of each separate population where the corresponding allele repeat size was observed in other sampled populations were considered. This measure reflects for each population the proportion of mutations present within the allele size range of microsatellites and low values are expected when alleles have been lost during previous bottleneck(s). Simulations assuming a constant mutation rate and TPM with 80–90% SMM have shown that M ratios smaller than 0.68 can be assumed to represent a loss of alleles during a previous reduction in population size (Garza and Williamson 2001). By comparison, empirical data from various species show that stable populations have M ratios above 0.82, while populations with a known population reduction have M ratios below 0.70 (Garza and Williamson 2001). Moreover, the allele size range of each microsatellite locus was compared among the populations and the effects of any differences on the M ratios were assessed. These ranges were compared with the Iberian, Serbian and Romanian populations in three and four loci, respectively. To assess the effects of sample size, a larger (n = 419) Norwegian sample was included (from Haanes et al. 2010), as well as a smaller sub-sample of the Iberian population (n = 20).
The demographic history of each population was assessed using a hierarchic Bayesian coalescent-based approach for the microsatellite data (Beaumont 1999; Storz and Beaumont 2002), as implemented in MSVAR 1.3 (http://www.rubic.rdg.ac.uk/~mab/). The method assumes a stepwise mutation model and an ancestral population of prior stable size that started to decline or increase exponentially until the present population size. Through the genealogical history of the sample, which looking back in time may be considered as a sequence of mutations and coalescent events, it uses MCMC to estimate four demographic parameters: average mutation rate across loci (μ), present effective population size (N 1), ancestral effective size (N 0) and time (years) since the onset of decline or expansion (T). Means and variances of prior parameter distributions are drawn from hyperpriors with separate means and variances, which were set wide to allow chains to evolve freely (Table 1). Different starting values for the mutation rate (μ) may have an effect on posterior distributions (Storz and Beaumont 2002) and an average μ of 103.5 was used both for priors and hyperpriors, reflecting a typical average among microsatellite loci (Dallas 1992; Weber and Wong 1993; Zhang and Hewitt 2003). To assess the effects of different starting values in the other parameters, runs with different hyperprior means of N 0 and T were applied to each population (runs 1–6, Table 1). Several runs were also run with similar priors, applying starting values that assumed a large ancestral effective size (N 0 = 10,000) and no change in population size (N 0 = N 1, run 7–11, Table 1). The generation time was set to 8 years (following Kruuk et al. 2002) and posterior estimates of T were expressed as numbers of years. To assess the effects of genetic structure and gene flow, the analyses were performed both for each population separately and then with all the sampled populations pooled together. Convergence was established from the Gelman–Rubenstein statistics R (<1.13, Gelman et al. 1995; Beaumont 1999). Chains were run for at least 2 × 109 iterations with a thinning value of 104.
Results
Genetic diversity and differentiation
None of the populations showed significant deviations from Hardy–Weinberg equilibrium in any of the microsatellite loci, except for BM5004 (p < 0.0001) in the Hungarian population and NVHRT73 in the Lithuanian population (p < 0.005). A total of 195 microsatellite alleles, including 70 private, were found among the populations. For the mtDNA control region a total of 26 haplotypes were observed. There were clear differences in the level of genetic variation among the red deer populations. The microsatellite data shows that the Norwegian and Swedish populations had explicitly lower values of allelic richness, number of private alleles and gene diversity compared with the other sampled populations (Table 2). The Serbian and Romanian populations (Feulner et al. 2004) had in four of the microsatellite loci by comparison allelic richness (mean: 6.0, SE: 0.1) and gene diversities (mean: 0.82, SE: 0.01) that were higher than in the Scandinavian populations but similar to the other populations. In three loci, also the Iberian population had a high allelic richness (mean, 7.0; SE, 0.5) and a high gene diversity (mean, 0.80; SE, 0.03). In terms of mtDNA sequence variability, the number and diversity (h) of haplotypes were high in the Scottish and Hungarian populations, intermediate in the Norwegian and Lithuanian populations while only one single and unique haplotype was observed in the Swedish population (Table 3). Nucleotide diversity and theta followed the same pattern. Tajima’s D was not significant in any populations (Table 3).
Among the sampled populations the global overall F st was 0.17 (SE, 0.02) and the global overall F is was 0.05 (SE, 0.02), across the 14 microsatellite loci. All pairwise F st values between these populations were significant, and the ones involving the Scandinavian populations were especially high (Table 4). F is values were moderate in the Hungarian and Scandinavian populations but low in the two others (Table 2). Within each separate population the overall F st (theta) from substructure was low, except in the Scandinavian populations where it was moderate, but where it involved low F is estimates (small f) when accounted for (Table 2). By comparison, in four of the loci the Serbian and Romanian populations had an overall F st value of 0.08 (SE, 0.03) and most had a high F is value (range, 0.00–0.51; mean, 0.30; SE, 0.05). Across three loci, the Iberian population had a F is value of −0.01 (SE, 0.05).
The FCA analysis (Fig. 2) showed that the Norwegian and Swedish populations formed clear separate clusters far away from the other populations, with axis I and II explaining 5.66% and 4.96% of the microsatellite variation, respectively. The other three populations were distributed along a third axis which explained 4.32% of the variation.
The results of the STRUCTURE analyses showed that a partitioning of the individual genetic variation into five genetic clusters was the most probable (P (K = 5 | D) >0.99). This was supported by higher levels of variance among runs with K > 5 and by a pronounced higher ∆K (>300) for K = 5 than for the other K values (∆K < 6; Fig. S1). The five genetic clusters correspond exactly to the five sampled populations and show low levels of admixture (Fig. 3). Genetic structure was not detected within any of the sampled populations but this is probably due to the low sample sizes and because ∆K reflects the main genetic structure rather than lower hierarchical structure.
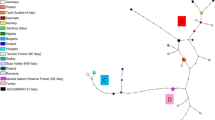
For the mtDNA sequences, the highest pairwise F st values involved the Hungarian population (Table 4), which represents the East European lineage (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). The Hungarian haplotypes were not observed in other populations and their distinctiveness was further evident from one to three inserts. Two of these were repeats in a sequence of similar nucleotides. The Hungarian haplotypes form a distinct clade that is supported by a mtDNA gene tree (not shown) and displayed by the network analysis (Fig. 4). As indicated by the haplotype diversities (h), the network showed that haplotype frequencies were more evenly distributed in the Scottish and Hungarian populations than in the Lithuanian and Norwegian populations. Among the populations, one haplotype was shared between the Lithuanian and the Scottish population but was only observed once in each population. Two haplotypes, one more common and one rare, were shared between the Norwegian (n = 7 and n = 1, respectively) and Scottish (n = 4 and n = 1, respectively) populations (Fig. 4).
Network analysis (TCS) based on the control region of mtDNA haplotypes found in red deer populations sampled in Hungary (Hu, brown), Lithuania (Li, green), Scotland (Sk, blue), Sweden (Sv, yellow) and Norway (No, red). Haplotype sizes are scaled by their observed frequency. Observed haplotypes are circular and haplotypes downloaded from GenBank are elongated. The dots represent missing intermediate haplotypes and the square is the haplotype assigned as most ancient by the network software (TCS)
Bottleneck signals and demographic histories
Among the 195 allele repeat sizes observed in our data set, each was observed in at least one population, but a total of 74, 96, 128, 132 and 75 were missing in either the Hungarian, Lithuanian, Swedish, Norwegian and Scottish population, respectively (Table S1). These gaps in the microsatellite repeat size range of each separate population were in other words present in one or more of the other populations. Increasing the Norwegian sample size by 20 times resulted in the detection of only 12 additional alleles across the 14 microsatellite loci. Further, within the allele size ranges of the 14 dinucleotide microsatellite loci, 50 repeat sizes representing potential mutations were not observed in any of the investigated populations. Among these potential mutations, 10 repeat sizes were observed for the four loci OarCP26, OarFCB304, BM888 and BM4208 in the Serbian and Romanian populations, together with 19 more repeat sizes outside the observed ranges of our data set. For the loci OarCP26 and OarFCB304, the additional repeats were also observed in the Iberian population. For the locus OarFCB193, two more of the repeat sizes missing within and two additional repeat sizes outside the ranges of the sampled populations were observed in the Iberian population (Table S1). With all these additional repeat sizes, these five loci showed near complete dinucleotide repeat size ranges across all the populations. However, across the four compared loci only 20–40 of totally 92 alleles were observed in each of the sampled Serbian and Romanian populations, involving several gaps in their repeat size ranges. By comparison, the Iberian population had nearly complete dinucleotide size ranges in two loci, but several gaps in the repeat size range of the third and less sampled locus. However, random sub-sampling of 20 Iberian red deer resulted in recording 8/13 and 9/12 of the alleles in the two loci with almost complete dinucleotide repeat size ranges, respectively.
A signal of a recent bottleneck of short duration or low severity was only detected in the Norwegian population. It was the only population with an excessive heterozygosity compared with what was expected given the observed number of alleles, or in other words which had an allele deficiency compared to what is expected from the observed heterozygosity (Table 5). The deficiency was statistically significant with a TPM mutation model of 80% SMM (p < 0.05) and nearly significant with 90% SMM (p < 0.07).
The observed M ratios were intermediate in the Hungarian population, relatively low in the Scottish and Lithuanian populations and especially low in the Swedish and Norwegian populations (Table 5). This reflects the proportion of alleles that were observed within the allele repeat size range of each population compared with the total number of alleles observed within this range across all the sampled populations. The observed allele ranges of microsatellite loci were narrower in the Scandinavian populations (Table S1), and if the repeat ranges observed in the other populations had been applied, their calculated M ratios would have been even lower (data not shown).
The hierarchical Bayesian analyses consistently supported population decline in all the investigated populations through estimates of large ancient (N 0) and small present (N 1) population sizes. In each of the populations, runs with different starting values for ancient population size or time since onset of decline (T) yielded similar posterior distributions of the parameters, as did also the several similar runs (Gelman–Rubensteins’ R <1.13). Among the populations, parameter estimates were similar for ancient population sizes (N 0 ∈ [50,000–100,000]) and times since onset of decline (T ∈ [5,000–10,000] years), but varied for the present populations sizes (N 1 ∈ [200–1,260]; Table 6). With all the populations pooled together an order of magnitude population decline was detected for the same time frame as for the separate populations (Table 6).
Discussion
The populations that were sampled for this study all have a history of postglacial colonisation. The Scandinavian, Scottish and Lithuanian populations have the same putative origin in the Iberian LGM refuge, while the Hungarian population has a putative origin in the Balkan LGM refuge (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). The lower levels of genetic variation observed in the Scandinavian populations for 14 microsatellite loci and one mtDNA sequence (Tables 2 and 3) thus suggest that genetic variation has been lost since postglacial colonisation, probably as a result of strong genetic drift during founder events or one or more subsequent bottlenecks. This was supported by the levels of genetic variation in the three and four loci compared with the Iberian and Serbian–Romanian populations, which lie within the two putative LGM refuges (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). The Iberian and Serbian–Romanian levels were also higher than the two Scandinavian but comparable with the other sampled populations, suggesting particular losses from the two Scandinavian populations. Genetic drift is also suggested by the genetic differentiation identified through FCA (Fig. 2) and pairwise F st values between the sampled populations (Table 4), especially in the Scandinavian populations. The high pairwise F st values and the STRUCTURE analyses suggest limited gene flow between and a low degree of admixture among the sampled populations.
Among the sampled populations, the missing alleles within microsatellite repeat size ranges could indicate previous bottleneck events but may also be biased by the sample sizes. However, in the Norwegian population the few additional alleles observed by increasing the sample size by 20 times (Haanes et al. 2010) suggest that the number of recorded alleles only was slightly affected by the sample size. The gaps within the microsatellite repeat size range of each separate population where the corresponding allele was observed in other sampled populations thus suggest that alleles have been lost during previous bottlenecks, especially in the Scandinavian populations. This is reflected by the M ratios. In all the sampled populations the same 50 microsatellite repeats were absent across the 14 loci, but the 12 of these repeats that were observed across five loci in the Iberian, Serbian and Romanian populations suggests that these gaps are related to demographic effects rather than complex multistep mutation. This is supported by the near complete dinucleotide repeat ranges in these five loci across all populations. In the Iberian, Serbian and Romanian populations, the additional 21 repeats observed in these five loci outside the observed ranges of the sampled populations provide further support for previous bottlenecks. However, the many gaps within the repeat size ranges of also the Serbian and Romanian populations (Table S1) may indicate that these populations have not been as demographically stable as presumed. By comparison, the sub-sampling in the two loci with almost complete dinucleotide repeat ranges of only a few Iberian individuals indicated that these recorded numbers of alleles were not much affected by sample size.
The allele deficiency in the Norwegian population indicates that alleles have been lost to a greater extent than heterozygosity, as would be expected from a recent bottleneck event of short duration or low severity (Maruyama and Fuerst 1985; Allendorf 1986; Cornuet and Luikart 1996). Such an event would concur well with the population size and timing of the population reduction recorded during the eighteenth and nineteenth centuries (Collett 1909; Ingebrigtsen 1924), and furthermore, the present genetic structure is concordant with the known distribution at that time (Haanes et al. 2010). A bottleneck of low severity does however not explain the reduced heterozygosity. Heterozygosity is only reduced during severe bottlenecks when common alleles are lost after the initial loss of rare alleles (Maruyama and Fuerst 1985; Allendorf 1986; Amos and Balmford 2001). Therefore, the reduced level in the Norwegian population suggests another older and more severe bottleneck event. By comparison, the recorded decline in the Swedish population from 300 to 50 years ago was more severe (Lønnberg 1906; Ahlèn 1965) and is compatible with its low observed genetic diversity. A loss of common alleles leading to reductions in both heterozygosity and allelic diversity could explain why no allele deficiency was observed after this recent population reduction. An alternative explanation is that an earlier bottleneck could have purged the rarest alleles so that the remaining and more common alleles were less sensitive to further allelic loss during a subsequent bottleneck. Moreover, the intermediate genetic structure within each of the Scandinavian populations (Table 2) may involve a bias in the BOTTLENECK analysis, where an increase in the number of rare alleles present in a population from immigration or population substructure can hide a bottleneck signal (Cornuet and Luikart 1996). Even stronger bottleneck signals have thus previously been detected for separate Norwegian localities through the BOTTLENECK analyses (Haanes et al. 2010).
The Bayesian analyses suggested ancient population reductions in all the sampled populations (Table 6). The time frames of 5,000–10,000 years suggested for the onset of decline in all the populations coincides with the period around postglacial colonisation (9,500–7,000 BP). After the last ice age, founder events and isolation after leading edge expansions involved loss of genetic variation and increased homozygosity in many newly colonised areas (Hewitt 2000; Hewitt 2004). Such losses during colonisation seem likely in areas like Scandinavia and Scotland, considering the long distance to and the short period of southward connection with mainland Europe (Andersen and Borns 1994), especially in the Norwegian population that has been isolated on the west coast of Norway (Ahlèn 1965). The different magnitudes of population decline suggested by the Bayesian analyses could be explained by founder events of different magnitudes during colonisation, which would also help explain the different M ratios.
However, the Bayesian estimates may also be biased from model assumptions. The mutation rate applied in the MSVAR analysis represents a typical average among microsatellites (Dallas 1992; Weber and Wong 1993; Zhang and Hewitt 2003). Slower mutation rates (10−6) yielded higher population sizes and much longer time frames (data not shown). Furthermore, the model assumes only a single change in population size and a constant population size prior to that (Beaumont 1999), and in populations with more than one previous bottleneck, as indicated in the Scandinavian populations, parameter estimates may be biased. Other assumptions are that mutations occur by SMM and an absence of population substructure (Goossens et al. 2006). For coalescent-based analyses such as MSVAR, genetic structure among the sampled populations may involve a bias in detecting bottleneck signals. When populations are interconnected by gene flow, false bottleneck signals can be created in separate populations when immigrant alleles with a long-term coalescent history are sampled along with resident alleles with a short-term coalescent history (Chikhi et al. 2010). By comparison, collapsing samples from genetically differentiated populations into one sample may give an opposite bias, hiding bottleneck signals. Considering the genetic structure, the low degree of admixture and the geographical distance between the sampled populations, the bias from immigrant non-resident gene copies should be of low magnitude in our MSVAR analyses. Non-indigenous gene copies may also arrive by translocation and introduction, but available studies indicate very limited impacts in the sampled populations (Gill 1990; Skog et al. 2009; Pérez-Espona et al. 2008; Haanes et al. 2010). Moreover, the pooling together of all the sampled populations in the MSVAR analysis involved detection of an order of magnitude population decline within the same time frame. The skewness in reproductive success and sex ratio associated with polygynous mating involve a pronounced reduction of the effective size of a population (Nunney 1993) and the parameter estimates of both ancient and current effective population sizes probably reflect much larger census sizes. The effective size of a population is also reduced when the sex ratio is skewed by male-biased harvest (Sæther et al. 2009). Since harvest is strongly male-biased in the Scandinavian countries compared with Scotland, Hungary and Eastern Europe (Milner et al. 2006), there might be a bias in the sex ratio among the sampled populations affecting their ratios between effective and census size. The Norwegian population currently has a very large census size (Langvatn 1988; Forchhammer et al. 1998; Langvatn 1998) and a highly skewed sex ratio could help explain its lower estimated current effective size.
The single mtDNA haplotype observed in the Swedish population was a strong indication of a long or severe bottleneck. However, the relatively similar levels of microsatellite variation in the two Scandinavian populations did not correspond to the higher level of mtDNA variation in the Norwegian population. Possible explanations may be male immigration into the Swedish population, which would increase microsatellite variation, and female immigration (or translocation) into the Norwegian population, which would increase mtDNA haplotype diversity. Non-indigenous red deer have been introduced in Sweden since 1950, but into localities well separated from the sanctuary of indigenous Swedish red deer in Scania (Ahlèn 1965; Gyllensten et al. 1983), which has remained relatively isolated (Gill 1990). The single observed Swedish haplotype suggests that female immigration is absent. Male red deer may, however, disperse as far as 200 km (Haanes unpublished data) and male immigration from locations where red deer were introduced could explain the higher microsatellite variation but low mtDNA variation in the indigenous Swedish population. This may also be the case in the Hungarian population, where the same pattern was observed.
A gene tree (not shown) and the mtDNA haplotype network did not support any other distinct clades other than for the Hungarian population, which represents the East European lineage (Ludt et al. 2004; Sommer et al. 2008; Skog et al. 2009). In accordance with these studies, the few mutations separating the haplotypes from Norway, Sweden, Lithuania and most Scottish haplotypes suggest a recent divergence and possibly a common origin of these populations in a glacial refuge prior to postglacial colonisation. The single unique haplotype in the Swedish population suggest both strong genetic drift and a limited gene flow between the Scandinavian populations. Archaeological finds also suggest separate prehistoric Scandinavian populations (Grieg 1908), but it should be noted that these distributions may have been wider since finds have been limited to localities with high pH and good conditions for conservation (Ahlèn 1965). By comparison, a high affinity is suggested between the Norwegian and Scottish populations by their sharing of two of the same haplotypes, as well as by their relatively low mtDNA-based F st value. During the Boreal period (9,000–7,500 BP), the channel between Norway and the North Sea land was at least 50 km wide at the most narrow places, and red deer migration across this channel from the British Isles seems unlikely (Grieg 1908; Hufthammer 2006). Still, we cannot rule out that human transport of red deer from Great Britain to Norway have occurred in earlier times like for example the Viking Age. Considering the higher suitability of females for such transport, this could explain the relatively high number of mtDNA haplotypes in Norway and the haplotype sharing with Scotland, as each introduced female may represent a separate matriline.
In conclusion, the Scandinavian populations seem to have lost genetic variation during one recent and one or several older population reductions. In the Norwegian population the recent bottleneck was of low severity but involved significant genetic drift. By comparison, the longer-lasting recent population reduction in Sweden was much more severe and may explain both loss of alleles and reduced heterozygosity, possibly overshadowing signals from other previous bottlenecks. The presently low level of mtDNA compared with microsatellite variation in indigenous Swedish red deer may indicate that non-indigenous male red deer have dispersed from introduced localities, which recently have increased in numbers (Gill 1990), and reproduced within the protected indigenous population. Managers should be aware and special actions considered for conservation of still-existing indigenous Swedish red deer. The single and uniquely observed haplotype may be used to monitor spatial distribution and expansion of indigenous Swedish red deer, as well as to detect introgression from female non-indigenous red deer. By comparison, the loss of genetic variation from the other sampled populations most probably derives from older and more severe or longer-lasting bottlenecks than the known recent decline in the eighteenth and nineteenth centuries, probably during founder events of postglacial colonisation in the early Holocene. Loss of genetic variation can affect the adaptive capacity of a population (Allendorf 1986; Soulé and Mills 1992; Nunney 2000), but this could not be assessed since we only used neutral genetic markers.
References
Ahlèn I (1965) Studies on the red deer, Cervus elaphus L. Scandinavia. III. Ecological investigations. Viltrevy 3:177–376
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Amos W, Balmford A (2001) When does conservation genetics matter? Heredity 87:257–265
Amos W, Harwood J (1998) Factors affecting levels of genetic diversity in natural populations. Phil Trans R Soc B 353:177–186
Andersen BG, Borns HW (1994) The ice age world. Scandinavian University Press, Oslo
Beaumont MA (1999) Detecting population expansion and decline using microsatellites. Genetics 153:2013–2029
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) GENETIX 4.05, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions,CNRS UMR 5171, Université de Montpellier II, Montpellier, FR.
Bhebhe E, Kogi J, Holder DA et al (1994) Caprine microsatellite dinucleotide repeat polymorphism at the SR-CRSP-6, SR-CRSP-7, SR-CRSP-8, SR-CRSP-9 and SR-CRSP-10. Anim Genet 25:203
Bishop MD, Kappes SM, Keele JW et al (1994) A genetic linkage map for cattle. Genetics 136:619–639
Buchanan FC, Crawford AM (1993) Ovine microsatellites at the OarFCB11, OarFCB128, OarFCB193, OarFCB226 and OarFCB304 loci. Anim Genet 24:145
Chikhi L, Sousa VC, Luisi P et al. (2010) The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186:983–995
Clark PU, Mix AC (2002) Ice sheets and sea level of the last glacial maximum. Quat Sci Rev 21:1–7
Clark PU, Dyke AS, Shakun JD et al (2009) The last glacial maximum. Science 325:710–714
Claussøn Friis P (1599) Om diur, fiske, fugle og træer udi Norrige (in Norwegian). In: Storm G (ed.) Samlede skrifter af peder claussøn friis. 1881. Brøgger, Kristiania, No
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Collett R (1909) Hjorten i Norge (Cervus elaphus atlanticus), nogle biologiske meddelelser (in Norwegian). Bergens mus Aarbok 6:9–31
Collett R (1912) Norges patterdyr (in Norwegian). H.Aschehoug & Co, Kristiania
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Dallas JF (1992) Estimation of microsatellite mutation rates in recombinant inbred strains of mouse. Mamm Genome 3:452
Di Rienzo A, Peterson AC, Garza JC et al (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci U S A 91:3166–3170
Ede AJ, Pierson CA, Crawford AM (1995) Ovine microsatellites at the OarCP9, OarCP16, OarCP20, OarCP21, OarCP23 and OarCP26 loci. Anim Genet 25:129–130
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver.3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Feulner PG, Bielfeldt W, Zachos FE et al (2004) Mitochondrial DNA and microsatellite analyses of the genetic status of the presumed subspecies Cervus elaphus montanus (Carpathian red deer). Heredity 93:299–306
Flagstad O, Røed KH (2003) Refugial origins of reindeer (Rangifer tarandus L.) inferred from mitochondrial DNA sequences. Evolution 57:658–670
Flerov KK (1952) Fauna of the U.S.S.R. USSR academy of sciences, Moscow
Forchhammer MC, Stenseth NC, Post E, Langvatn R (1998) Population dynamics of Norwegian red deer: density-dependence and climatic variation. Proc R Soc B 265:341–350
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Gelman A, Carlin JB, Stern HS, Rubin DB (1995) Bayesian data analysis. Chapman and Hall, London
Gill R (1990) Monitoring the status of European and North American Cervids. In: The global environment monitoring system information series. United Nations environment programme, Nairobi
Goossens B, Chikhi L, Ancrenaz M et al (2006) Genetic signature of anthropogenic population collapse in orang-utans. PLoS Biol 4:285–291
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices. Available from http://www.unil.ch/izea/softwares/fstat.html
Grieg JA (1908) Bidrag til kunnskaben om Norges hvirveldyrsfauna i ældre tider (in Norwegian). Bergens mus Aarbok 7:1–34
Grieg JA (1909) Hjortens utbredelse i Norge i forhistorisk tid (in Norwegian). Naturen 33:65–78
Gyllensten U, Ryman N, Reuterwall C, Dratch P (1983) Genetic differentiation in four European subspecies of red deer (Cervus elaphus L.). Heredity 51:561–580
Haanes H, Røed KH, Flagstad Ø, Rosef O (2010) Genetic structure in an expanding cervid population after population reduction. Conserv Genet 11:11–20. doi:10.1007/s10592-008-9781-0
Haanes H, Røed KH, Mysterud A, et al. (2010) Consequences for genetic diversity and population performance of introducing continental red deer into the northern distribution range. Conserv Genet 11:1653–1665. doi:10.1007/s10592-010-0048-1
Hartl GB, Willing R, Lang G, Klein F, Koller J (1990) Genetic variability and differentiation in red deer (Cervus elaphus L) of Central-Europe. Genet Sel Evol 22:289–306
Hartl GB, Lang G, Klein F, Willing R (1991) Relationship between allozymes, heterozygosity and morphological characters in red deer (Cervus elaphus), and the influence of selective hunting on allele frequency distribution. Heredity 66:343–350
Hartl GB, Zachos F, Nadlinger K (2003) Genetic diversity in European red deer (Cervus elaphus L.): anthropogenic influences on natural populations. CR Biol 326:37–42
Hartl GB, Zachos FE, Nadlinger K et al (2005) Allozyme and mitochondrial DNA analysis of French red deer (Cervus elaphus) populations: genetic structure and its implications for management and conservation. Mamm Biol 70:24–34
Herzog S, Gehle T (2001) Genetic structures and clinal variation of European red deer Cervus elaphus populations for two polymorphic gene loci. Wildl Biol 7:55–59
Hewitt GM (1996) Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc 58:247–276
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hewitt GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Phil Trans R Soc Lond B 359:183–195
Hufthammer AK (2006) The vertebrate fauna of eastern Norway—from the ice age to the middle ages. Skrifter (Kulturhistorisk Museum, Oslo) 4:191–202
Hulme DJ, Silk JP, Redwin JM, Barendse W, Beh KJ (1994) Ten polymorphic ovine microsatellites. Anim Genet 25:434–435
Ingebrigtsen O (1924) Hjortens utbredelse i Norge (in Norwegian). Bergens Museums Aarbok 1922–1923 Naturvitensk. Række 6:1–58
Jonsson L (1995) Vertebrate fauna during the Mesolithic on the Swedish west coast. In: Fischer A (ed) Man and sea in Mesolithic. Oxbow books, Oxford, pp 147–160
Kimura M, Ohta T (1978) Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci U S A 75:2868–2872
Kruuk LEB, Slate J, Pemberton JM et al (2002) Antler size in red deer: heritability and selection but no evolution. Evolution 56:1683–1695
Kuehn R, Schroeder W, Pirchner F, Rottmann O (2003) Genetic diversity, gene flow and drift in Bavarian red deer populations (Cervus elaphus). Conserv Genet 4:157–166
Kuehn R, Haller H, Schroeder W, Rottmann O (2004) Genetic roots of the red deer (Cervus elaphus) population in eastern Switzerland. J Hered 95:136–143
Kurtèn B (1968) Pleistoscene mammals of Europe. Weidenfeld and Nicholson, London
Langvatn R (1988) Hjortens utbredelse i Norge-en oversigt (in Norwegian). Villreinen 1:1–8
Langvatn R (1998) Hjortens erobring av Norge (in Norwegian). In: Brox KH (ed) Brennpunkt natur. Tapir cop, Trondheim, pp 49–71
Lavsund S (1975) Kronhjortens, Cervus elaphus L., utbredning i Sverige 1900–1973 (in Swedish). Rapporter och uppsatser for Inst för Skogszoologi, Skogshögskolan, Stockholm 18:19–27
Lavsund S (1990) Kronhjorten i Sverige och dess framtid (in Swedish). Viltnytt 28:18–26
Liljegren R, Ekström J (1996) The terrestrial late glacial fauna in South Sweden. Acta Archaeol Lund Ser 8 24:135–139
Lister AM (1984) Evolutionary and ecological origins of British deer. Proc R Soc Edinb B 82:205–229
Lister AM (1993) The stratigraphical significance of deer species in the cromer forest-bed formation. J Quat Sci 8:95–108
Lønnberg E (1906) On the geographic races of red deer in Scandinavia. Arch Zool 3:1–19
Ludt CJ, Schroeder W, Rottmann O, Kuehn R (2004) Mitochondrial DNA phylogeography of red deer (Cervus elaphus). Mol Phylogenet Evol 31:1064–1083
Maruyama T, Fuerst PA (1985) Population bottlenecks and nonequilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
Milner JM, Bonenfant C, Mysterud A et al (2006) Temporal and spatial development of red deer harvesting in Europe: biological and cultural factors. J Appl Ecol 43:721–734
Moore SS, Byrne K, Berger KT et al (1994) Characterization of 65 bovine microsatellites. Mamm Genome 5:84–90
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M et al (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nunney L (1993) The influence of mating system and overlapping generations on effective population size. Evolution 47:1329–1341
Nunney L (2000) The value of effective population size. In: Clegg MT, Hecht MK, MacIntyre RJ (eds) The limits to knowledge in conservation genetics. Kluwer Academic, New York
Nussey DH, Pemberton J, Donald A, Kruuk LE (2006) Genetic consequences of human management in an island population of red deer (Cervus elaphus). Heredity 97:56–65
Nystrom V, Angerbjorn A, Dalen L (2006) Genetic consequences of a demographic bottleneck in the Scandinavian arctic fox. Oikos 114:84–94
Pérez-Espona S, Pérez-Barbeíra FJ, McLeod JE et al (2008) Landscape features affect gene flow of Scottish highland red deer (Cervus elaphus). Mol Ecol 17:981–996
Peréz-González J, Carranza J, Torres-Porras J, Fernández-García JL (2010) Low heterozygosity at microsatellite markers in Iberian red deer with small antlers. J Hered 101:553–561
Pontoppidan (1753) Norges naturlige historie (in Norwegian). Roskilde og Bagger, København, DK
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Randi E, Mucci N, Claro-Hergueta F et al (2001) A mitochondrial DNA control region phylogeny of the Cervinae: speciation in Cervus and implications for conservation. Anim Conserv 4:1–11
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Røed KH (1998) Microsatellite variation in Scandinavian Cervidae using primers derived from Bovidae. Hereditas 129:19–25
Røed KH, Midthjell L (1998) Microsatellites in reindeer, Rangifer tarandus, and their use in other cervids. Mol Ecol 7:1773–1778
Rosvold J (2006) Do single prehistoric human dvelling sites reveal changes in the relative abundance of moose and red deer in Western Norway? Master thesis, Norwegian University of Science and Technology
Rousset F, Raymond M (1995) Testing heterozygote excess and deficiency. Genetics 140:1413–1419
Sæther BE, Engen S, Solberg EJ (2009) Effective size of harvested ungulate populations. Anim Conserv 12:488–495
Skog A, Zachos FE, Rueness EK et al (2009) Phylogeography of red deer (Cervus elaphus) in Europe. J Biogeogr 36:66–77
Sommer RS, Nadachowski A (2006) Glacial refugia of mammals in Europe: evidence from fossil records. Mamm Rev 36:251–265
Sommer RS, Zachos FE, Street M et al (2008) Late Quaternary distribution dynamics and phylogeography of the red deer (Cervus elaphus) in Europe. Quat Sci Rev 27:714–733
Soulé ME, Mills S (1992) Conservation genetics and conservation biology: a troubled marriage. In: Sandlund OT, Hindar K, Brown AHD (eds) Conservation of biodiversity for sustainable development. Scandinavian University Press, Oslo
Storz JF, Beaumont MA (2002) Testing for genetic evidence of population expansion and contraction: an empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56:154–166
Strandgaard H, Simonsen V (1993) Genetic differentiation in population of red deer, Cervus elaphus, in Denmark. Hereditas 119:171–177
Vila C, Sundqvist AK, Flagstad O et al (2003) Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc R Soc B 270:91–97
Weber JL, Wong C (1993) Mutation of human short tandem repeats. Hum Mol Genet 2:1123–1128
Weber DS, Stewart BS, Garza JC, Lehman N (2000) An empirical genetic assessment of the severity of the northern elephant seal population bottleneck. Curr Biol 10:1287–1290
Weber DS, Stewart BS, Lehman N (2004) Genetic consequences of a severe population bottleneck in the Guadalupe fur seal (Arctocephalus townsendi). J Hered 95:144–153
Weir BS (1996) Genetic data analysis II: methods for discrete population genetic data. Sinauer Associates, Sunderland
Whalstöm K (1996) Förvaltning av den svenska kronhjortsstammen; skötsel, viltskade-, och rasproblematik (in Swedish). Report from Svenska Jägareförbundet, Forskningsavdelningen, Uppsala, Sweeden
Wilson GA, Strobeck C, Wu L, Coffin J (1997) Characterization of microsatellite loci in caribou Rangifer tarandus, and their use in other artiodactyls. Mol Ecol 6:697–699
Zachos F, Hartl GB, Apollonio M, Reutershan T (2003) On the phylogeographic origin of the Corsican red deer (Cervus elaphus corsicanus): evidence from microsatellites and mitochondrial DNA. Mamm Biol 68:284–298
Zhang DX, Hewitt GM (2003) Nuclear DNA analyses in genetic studies of populations: practice, problems and prospects. Mol Ecol 12:563–584
Acknowledgements
For providing samples, we thank Dr. Aligmantas Paulauskas at the Vytautas Magnus University in Lithuania, Dr. László Szemethy at the St. Stephen University in Hungary, Per Larsson and Göran Anderson. For kindly providing microsatellite data from Serbia and Romania, we are in debt to and thank Dr. Frank Zachos at the Christian-Albrechts University, Kiel, Germany. For kindly providing microsatellite data from an Iberian population, we are in debt to and thank Dr. Javier Pérez-González at the University of Extremadura, Cáceres, Spain. Information about the Swedish population was kindly provided by Björn Molitor. For help in the laboratory, we are in debt to Liv Midthjell and for helpful assistance with MSVAR analyses and loan of large computational power we thank Kjetil Olsen and the IT group at the Norwegian School of Veterinary Science. For helpful comments and improvements of the manuscript, we thank Anders Jarnemo, at the Swedish University of Agricultural Sciences in Sweden, as well as two anonymous reviewers. For help with native proof reading, we thank Associate Professor David Griffiths, Institute of Basic Sciences and Aquatic Medicine at the Norwegian School of Veterinary Sciences.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Alves
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Posterior probability (ln P(D | K); mean (−) and range of 2 SDs) and ∆K (o) for a data set of 419 Norwegian red deer, given different numbers of genetic cluster (K ∈ [1, 7]), each run 10 times for an admixture model with correlated gene frequencies in STRUCTURE. (DOC 105 kb)
Table S1
Comparison of the microsatellite mutation repeat sizes and ranges, observed among European red deer populations in Scotland (Sc), Norway (No), Sweden (Sw), Lithuania (Li) and Hungary (Hu) across 14 loci (n ∈ [19, 20]) . These are compared, (1) with the kind permission of Dr. Frank Zachos in four loci with nine populations (n ∈ [6, 26]) in Serbia (BAC, BAO and BAF) and Romania (Feulner et al. 2004), and (2) with the kind permission Dr. Javi Peréz-González in three loci with one population (SP; OarCP26; n = 27, OarFCB304 and OarFCB193; n = 240) in Iberia (Peréz-González et al. 2010). Allele sizes are given in absolute numbers of bases and the populations they are observed are marked X. To assess sample size effects, a larger Norwegian sample (NN; n = 419, Haanes et al. 2010) and a smaller Spanish sub-sample in two loci (S1; n = 20) are included in the table. (DOC 1139 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Haanes, H., Røed, K.H., Perez-Espona, S. et al. Low genetic variation support bottlenecks in Scandinavian red deer. Eur J Wildl Res 57, 1137–1150 (2011). https://doi.org/10.1007/s10344-011-0527-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10344-011-0527-9