Abstract
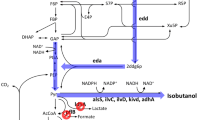
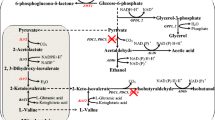
Isobutanol as a more desirable biofuel has attracted much attention. In our previous work, an isobutanol-producing strain Escherichia coli LA09 had been obtained by rational redox status improvement under guidance of the genome-scale metabolic model. However, the low transformation from sugar to isobutanol is a limiting factor for isobutanol production by E. coli LA09. In this study, the intracellular metabolic profiles of the isobutanol-producing E. coli LA09 with different initial glucose concentrations were investigated and the metabolic reaction of fructose 6-phosphate to 1, 6-diphosphate fructose in glycolytic pathway was identified as the rate-limiting step of glucose transformation. Thus, redesigned carbon catabolism was implemented by altering flux of sugar metabolism. Here, the heterologous Entner–Doudoroff (ED) pathway from Zymomonas mobilis was constructed, and the adaptation of upper and lower parts of ED pathway was further improved with artificial promoters to alleviate the accumulation of toxic intermediate metabolite 2-keto-3-deoxy-6-phospho-gluconate (KDPG). Finally, the best isobutanol-producing E. coli ED02 with higher glucose transformation and isobutanol production was obtained. In the fermentation of strain E. coli ED02 with 45 g/L initial glucose, the isobutanol titer, yield and average producing rate were, respectively, increased by 56.8, 47.4 and 88.1% to 13.67 g/L, 0.50 C-mol/C-mol and 0.456 g/(L × h) in a shorter time of 30 h, compared with that of the starting strain E. coli LA09.






Similar content being viewed by others
References
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86-U13. https://doi.org/10.1038/nature06450
Baez A, Cho K-M, Liao JC (2011) High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl Microbiol Biotechnol 90:1681–1690. https://doi.org/10.1007/s00253-011-3173-y
Bastian S, Liu X, Meyerowitz JT, Snow CD, Chen MMY, Arnold FH (2011) Engineered ketol-acid reductoisomerase and alcohol dehydrogenase enable anaerobic 2-methylpropan-1-ol production at theoretical yield in Escherichia coli. Metab Eng 13:345–352. https://doi.org/10.1016/j.ymben.2011.02.004
Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, Eikmanns BJ (2011) Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol 77:3300–3310. https://doi.org/10.1128/aem.02972-10
Buijs NA, Siewers V, Nielsen J (2013) Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr Opin Chem Biol 17:480–488. https://doi.org/10.1016/j.cbpa.2013.03.036
Chong L (2001) Molecular cloning—a laboratory manual, 3rd edition. Science 292:446. https://doi.org/10.1126/science.1060677
Conway T, Fliege R, Jones-Kilpatrick D, Liu J, Barnell WO, Egan SE (1991) Cloning, characterization and expression of the Zymononas mobilis eda gene that encodes 2-keto-3-deoxy-6-phosphogluconate aldolase of the Entner–Doudoroff pathway. Mol Microbiol 5:2901–2911. https://doi.org/10.1111/j.1365-2958.1991.tb01850.x
Felpeto-Santero C, Rojas A, Tortajada M, Galan B, Ramon D, Garcia JL (2015) Engineering alternative isobutanol production platforms. AMB Express. https://doi.org/10.1186/s13568-015-0119-2
Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R (2013) Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci USA 110:10039–10044. https://doi.org/10.1073/pnas.1215283110
Fu J, Wang Z, Chen T, Liu W, Shi T, Wang G, Y-j Tang, Zhao X (2014) NADH plays the vital role for chiral pure D-(-)-2,3-butanediol production in bacillus subtilis under limited oxygen conditions. Biotechnol Bioeng 111:2126–2131. https://doi.org/10.1002/bit.25265
Gaida SM, Liedtke A, Jentges AHW, Engels B, Jennewein S (2016) Metabolic engineering of Clostridium cellulolyticum for the production of n-butanol from crystalline cellulose. Microb Cell Fact. https://doi.org/10.1186/s12934-015-0406-2
Hao T, Han B, Ma H, Fu J, Wang H, Wang Z, Tang B, Chen T, Zhao X (2013) In silico metabolic engineering of Bacillus subtilis for improved production of riboflavin, Egl-237, (R, R)-2,3-butanediol and isobutanol. Mol BioSyst 9:2034–2044. https://doi.org/10.1039/c3mb25568a
Hauf J, Zimmermann FK, Muller S (2000) Simultaneous genomic overexpression of seven glycolytic enzymes in the yeast Saccharomyces cerevisiae. Enzym Microb Technol 26:688–698. https://doi.org/10.1016/s0141-0229(00)00160-5
Heckman KL, Pease LR (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc 2:924–932. https://doi.org/10.1038/nprot.2007.132
Huerta-Beristain G, Utrilla J, Hernandez-Chavez G, Bolivar F, Gosset G, Martinez A (2008) Specific ethanol production rate in ethanologenic Escherichia coli strain KO11 is limited by pyruvate decarboxylase. J Mol Microbiol Biotechnol 15:55–64. https://doi.org/10.1159/000111993
Jojima T, Inui M (2015) Engineering the glycolytic pathway: a potential approach for improvement of biocatalyst performance. Bioengineered 6:328–334. https://doi.org/10.1080/21655979.2015.1111493
Kalnenieks U (2006) Physiology of Zymomonas mobilis some unanswered questions. In: Poole RK (ed) Advances in microbial physiology, vol 51. Academic Press, Cambridge, pp 73–117. https://doi.org/10.1016/S0065-2911(06)51002-1
Kandror O, DeLeon A, Goldberg AL (2002) Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci USA 99:9727–9732. https://doi.org/10.1073/pnas.142314099
Karabektas M, Hosoz M (2009) Performance and emission characteristics of a diesel engine using isobutanol-diesel fuel blends. Renew Energy 34:1554–1559. https://doi.org/10.1016/j.renene.2008.11.003
Kim J, Jeon CO, Park W (2008) Dual regulation of zwf-1 by both 2-keto-3-deoxy-6-phosphogluconate and oxidative stress in Pseudomonas putida. Microbiology SGM 154:3905–3916. https://doi.org/10.1099/mic.0.2008/020362-0
Kolodziej R, Scheib J (2012) Bio-isobutanol: the next-generation biofuel. Hydrocarb Processing 91:79+
Lara AR, Caspeta L, Gosset G, Bolivar F, Ramirez OT (2008) Utility of an Escherichia coli strain engineered in the substrate uptake system for improved culture performance at high glucose and cell concentrations: an fed-batch cultures. Biotechnol Bioeng 99:893–901. https://doi.org/10.1002/bit.21664
Li S, Huang D, Li Y, Wen J, Jia X (2012) Rational improvement of the engineered isobutanol-producing Bacillus subtilis by elementary mode analysis. Microb Cell Fact. https://doi.org/10.1186/1475-2859-11-101
Li S, Wen J, Jia X (2011) Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl Microbiol Biotechnol 91:577–589. https://doi.org/10.1007/s00253-011-3280-9
Li S, Xu N, Liu L, Chen J (2014) Engineering of carboligase activity reaction in Candida glabrata for acetoin production. Metab Eng 22:32–39. https://doi.org/10.1016/j.ymben.2013.12.005
Lin Z, Xu Z, Li Y, Wang Z, Chen T, Zhao X (2014) Metabolic engineering of Escherichia coli for the production of riboflavin. Microb Cell Fact. https://doi.org/10.1186/s12934-014-0104-5
Liu J, Qi H, Wang C, Wen J (2015) Model-driven intracellular redox status modulation for increasing isobutanol production in Escherichia coli. Biotechnol Biofuels. https://doi.org/10.1186/s13068-015-0291-2
Liu Y, Zhou H, Wang L, Wang S, Fan L (2016) Improving Saccharomyces cerevisiae growth against lignocellulose-derived inhibitors as well as maximizing ethanol production by a combination proposal of gamma-irradiation pretreatment with in situ detoxification. Chem Eng J 287:302–312. https://doi.org/10.1016/j.cej.2015.10.086
Lu J, Brigham CJ, Gai CS, Sinskey AJ (2012) Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl Microbiol Biotechnol 96:283–297. https://doi.org/10.1007/s00253-012-4320-9
Ng CY, Farasat I, Maranas CD, Salis HM (2015) Rational design of a synthetic Entner–Doudoroff pathway for improved and controllable NADPH regeneration. Metab Eng 29:86–96. https://doi.org/10.1016/j.ymben.2015.03.001
Nie L, Wu G, Culley DE, Scholten JCM, Zhang W (2007) Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit Rev Biotechnol 27:63–75. https://doi.org/10.1080/07388550701334212
Perrenoud A, Sauer U (2005) Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187:3171–3179. https://doi.org/10.1128/jb.187.9.3171-3179.2005
Purvis JE, Yomano LP, Ingram LO (2005) Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl Environ Microbiol 71:3761–3769. https://doi.org/10.1128/aem.71.7.3761-3769.2005
Qi H, Li S, Zhao S, Huang D, Xia M, Wen J (2014) Model-driven redox pathway manipulation for improved isobutanol production in Bacillus subtilis complemented with experimental validation and metabolic profiling analysis. PLoS One. https://doi.org/10.1371/journal.pone.0093815
Rogers PL, Jeon YJ, Lee KJ, Lawford HG (2007) Zymomonas mobilis for fuel ethanol and higher value products. In: Olsson L (ed) Biofuels, vol 108. Advances in biochemical engineering-biotechnology. Springer, Berlin, Heidelberg, pp 263–288. https://doi.org/10.1007/10_2007_060
Schaaff I, Heinisch J, Zimmermann FK (1989) Overproduction of glycolytic enzymes in yeast. Yeast (Chichester, Engl) 5:285–290. https://doi.org/10.1002/yea.320050408
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C-T method. Nat Protoc 3:1101–1108. https://doi.org/10.1038/nprot.2008.73
Schuetz R, Zamboni N, Zampieri M, Heinemann M, Sauer U (2012) Multidimensional optimality of microbial metabolism. Science 336:601–604. https://doi.org/10.1126/science.1216882
Shi A, Zhu X, Lu J, Zhang X, Ma Y (2013) Activating transhydrogenase and NAD kinase in combination for improving isobutanol production. Metab Eng 16:1–10. https://doi.org/10.1016/j.ymben.2012.11.008
Smith KM, Cho K-M, Liao JC (2010) Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol 87:1045–1055. https://doi.org/10.1007/s00253-010-2522-6
Snoep JL, Yomano LP, Westerhoff HV, Ingram LO (1995) Protein burden in Zymomonas mobilis: negative flux and growth control due to overproduction of glycolytic enzymes. Microbiology 141:2329–2337
Wang B-W, Shi A-Q, Tu R, Zhang X-L, Wang Q-H, Bai F-W (2012) Branched-chain higher alcohols. In: Bai FW, Liu CG, Huang H, Tsao GT (eds) Biotechnology in China III: biofuels and bioenergy, vol 128. Advances in biochemical engineering-biotechnology. Springer, Berlin, Heidelberg, pp 101–118. https://doi.org/10.1007/10_2011_121
Wang C, Liu J, Liu H, Liang S, Wen J (2017) Combining metabolomics and network analysis to improve tacrolimus production in Streptomyces tsukubaensis using different exogenous feedings. J Ind Microbiol Biotechnol 44:1527–1540. https://doi.org/10.1007/s10295-017-1974-4
Xia M, Huang D, Li S, Wen J, Jia X, Chen Y (2013) Enhanced FK506 production in Streptomyces tsukubaensis by rational feeding strategies based on comparative metabolic profiling analysis. Biotechnol Bioeng 110:2717–2730. https://doi.org/10.1002/bit.24941
Zhang J, Fu R-Y, Hugenholtz J, Li Y, Chen J (2007) Glutathione protects Lactococcus lactis against acid stress. Appl Environ Microbiol 73:5268–5275. https://doi.org/10.1128/aem.02787-06
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21676189), the key technologies R & D program of Tianjin (No. 16YFZCSY00780).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, S., Chen, H., Liu, J. et al. Rational design of a synthetic Entner–Doudoroff pathway for enhancing glucose transformation to isobutanol in Escherichia coli. J Ind Microbiol Biotechnol 45, 187–199 (2018). https://doi.org/10.1007/s10295-018-2017-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-018-2017-5