Abstract
Fermentative processes are widely used to produce food, beverages and biofuels. Saccharomyces cerevisiae is an efficient ethanol-producing microorganism. However, a concentration of high ethanol and other metabolites can affect yeast viability and decrease the ethanol yield. Many studies have focused on improving the fermentative efficiency, mostly through the genetic engineering of genes that have a direct impact on specific metabolic pathways. In the present study, we characterized a small open reading frame encoding a protein with an unknown function and biological role termed YNR034W-A. We analyzed the expression profile of the YNR034W-A gene during growth and glucose treatment, finding that it is expressed during the diauxic shift and stationary phase and is negatively regulated by glucose. We overexpressed the YNR034W-A gene in the BY4741 laboratory strain and a wild-type yeast strain (AR5) isolated during the Tequila fermentation process. Transformant derivatives of the AR5 strain showed an improved fermentative efficiency during fermentation of Agave tequilana Weber juice. We suggest that the improved fermentative efficiency is the result of a higher stress tolerance response in the YNR034W-A overexpressing transformant.







Similar content being viewed by others
References
Aguilar-Uscanga B, Arrizon J, Ramirez J, Solis-Pacheco J (2007) Effect of Agave tequilana juice on cell wall polysaccharides of three Saccharomyces cerevisiae strains from different origins. Antonie Van Leeuwenhoek 91(2):151–157
Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314(5805):1565–1568
Argueso JL, Carazzolle MF, Mieczkowski PA et al (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res 19(12):2258–2270
Arrizon J, Gschaedler A (2007) Effects of the addition of different nitrogen sources in the tequila fermentation process at high sugar concentration. J Appl Microbiol 102(4):1123–1131
Arrizon J, Fiore C, Acosta G, Romano P, Gschaedler A (2006) Fermentation behaviour and volatile compound production by agave and grape must yeasts in high sugar Agave tequilana and grape must fermentations. Antonie Van Leeuwenhoek 89(1):181–189
Bai FW, Chen LJ, Zhang Z, Anderson WA, Moo-Young M (2004) Continuous ethanol production and evaluation of yeast cell lysis and viability loss under very high gravity medium conditions. J Biotechnol 110(3):287–293
Basrai MA, Hieter P, Boeke JD (1997) Small open reading frames: beautiful needles in the haystack. Genome Res 7(8):768–771
Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M (1998) Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol 180(5):1044–1052
Buck TM, Jordan R, Lyons-Weiler J, Adelman JL, Needham PG, Kleyman TR, Brodsky JL (2015) Expression of three topologically distinct membrane proteins elicits unique stress response pathways in the yeast Saccharomyces cerevisiae. Physiol Genomics 47(6):198–214
Cedeño MC (1995) Tequila Production. Crit Rev Biotechnol 15(1):1–11
Chen Y, Sheng J, Jiang T, Stevens J, Feng X, Wei N (2016) Transcriptional profiling reveals molecular basis and novel genetic targets for improved resistance to multiple fermentation inhibitors in Saccharomyces cerevisiae. Biotechnol Biofuels 9(1):1–18
Cheng C, Zhang M, Xue C, Bai F, Zhao X (2016) Development of stress tolerant Saccharomyces cerevisiae strains by metabolic engineering: new aspects from cell flocculation and zinc supplementation. J Biosci Bioeng. doi:10.1016/j.jbiosc.2016.07.021
Eisenberg T, Schroeder S, Andryushkova A et al (2014) Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab 19(3):431–444
Garay-Arroyo A, Covarrubias AA, Clark I, Nino I, Gosset G, Martinez A (2004) Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol 63(6):734–741
Gutiérrez-Lomelí M, Torres-Guzman JC, González-Hernández GA, Cira-Chávez LA, Pelayo-Ortiz C, de Ramírez-Córdova JJ (2008) Overexpression of ADH1 and HXT1 genes in the yeast Saccharomyces cerevisiae improves the fermentative efficiency during tequila elaboration. Antonie Van Leeuwenhoek 93(4):363–371
Hou L, Cao X, Wang C, Lu M (2009) Effect of overexpression of transcription factors on the fermentation properties of Saccharomyces cerevisiae industrial strains. Lett Appl Microbiol 49(1):14–19
Kim J, Kim E (2016) Rag GTPase in amino acid signaling. Amino Acids 48(4):915–928
Kobayashi N, McEnte K (1993) Identification of cis and trans components of a novel heat-shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol 13(1):248–256
Lachance MA (1995) Yeast communities in a natural tequila fermentation. Antonie Van Leeuwenhoek 68(2):151–160
Lai L-C, Kosorukoff AL, Burke PV, Kwast KE (2005) Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol Cell Biol 25(10):4075–4091
Lappe-Oliveras P, Moreno-Terrazas R, Arrizón-Gaviño J, Herrera-Suárez T, García-Mendoza A, Gschaedler-Mathis A (2008) Yeasts associated with the production of Mexican alcoholic nondistilled and distilled Agave beverages. FEMS Yeast Res 8(7):1037–1052
Levine TP, Daniels RD, Wong LH, Gatta AT, Gerondopoulos A, Barr FA (2014) Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4(2):62–69
Ma M, Liu ZL (2010) Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 87(3):829–845
Mancilla-Margalli NA, López MG (2002) Generation of Maillard compounds from inulin during the thermal processing of Agave tequilana Weber Var. azul. J Agric Food Chem 50(4):806–812
Marques AC, Vinckenbosch N, Brawand D, Kaessmann H (2008) Functional diversification of duplicate genes through subcellular adaptation of encoded proteins. Genome Biol 9(3):R54
Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15(9):2227–2235
Miller GL (1959) Use of Dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Mumber D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156(1):119–122
Murphy JP, Stepanova E, Everley RA, Paulo JA, Gygi SP (2015) Comprehensive temporal protein dynamics during the diauxic shift in Saccharomyces cerevisiae. Mol Cell Proteomics 14(9):2454–2465
Nagodawithana TW, Steinkraus KH (1976) Influence of the rate of ethanol production and accumulation on the viability of Saccharomyces cerevisiae in “rapid fermentation”. Appl Environ Microbiol 31(2):158–162
Olivas WM, Muhlrad D, Parker R (1997) Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res 25(22):4619–4625
Pais TM, Foulquié-Moreno MR, Hubmann G, Duitama J, Swinnen S, Goovaerts A, Yang Y, Dumortier F, Thevelein JM (2013) Comparative polygenic analysis of maximal ethanol accumulation capacity and tolerance to high ethanol levels of cell proliferation in yeast. PLoS Genet 9(6):e1003548–18
Palmqvist E, Grage H, Meinander NQ, Hahn-Hagerdal B (1999) Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng 63(1):46–55
Park SH, Koh SS, Chun JH, Hwang HJ, Kang HS (1999) Nrg1 is a transcriptional repressor for glucose repression of STA1 gene expression in Saccharomyces cerevisiae. Mol Cell Biol 19(3):2044–2050
Peter Smits H, Hauf J, Müller S, Hobley TJ, Zimmermann FK, Hahn-Hägerdal B, Nielsen J, Olsson L (2000) Simultaneous overexpression of enzymes of the lower part of glycolysis can enhance the fermentative capacity of Saccharomyces cerevisiae. Yeast 16(14):1325–1334
Powis K, Zhang T, Panchaud N, Wang R, De Virgilio C, Ding J (2015) Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res 25(9):1043–1059
Ramirez-Córdova J, Drnevich J, Madrigal-Pulido JA, Arrizon J, Allen K, Martínez-Velázquez M, Alvarez-Maya I (2012) Transcriptome analysis identifies genes involved in ethanol response of Saccharomyces cerevisiae in Agave tequilana juice. Antonie Van Leeuwenhoek 102(2):247–255
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141(2):290–303
Sasano Y, Watanabe D, Ukibe K, Inai T, Ohtsu I, Shimoi H, Takagi H (2012) Overexpression of the yeast transcription activator Msn2 confers furfural resistance and increases the initial fermentation rate in ethanol production. J Biosci Bioeng 113(4):451–455
Schaaff I, Heinisch J, Zimmermann FK (1989) Overproduction of glycolytic-enzymes in Yeast. Yeast 5(4):285–290
Sheff MA, Thorn KS (2004) Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21(8):661–670
Shi D-J, Wang C-L, Wang K-M (2008) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36(1):139–147
Smits HP, Hauf J, Muller S, Hobley TJ, Zimmermann FK, Hahn-Hagerdal B, Nielsen J, Olsson L (2000) Simultaneous overexpression of enzymes of the lower part of glycolysis can enhance the fermentative capacity of Saccharomyces cerevisiae. Yeast 16(14):1325–1334
Snoek T, Verstrepen KJ, Voordeckers K (2016) How do yeast cells become tolerant to high ethanol concentrations? Curr Genet 62(3):475–480
Taherzadeh MJ, Gustafsson L, Niklasson C, Lidén G (2000) Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl Microbiol Biotechnol 53(6):701–708
Tao X, Zheng D, Liu T, Wang P, Zhao W, Zhu M, Jiang X, Zhao Y, Wu X (2012) A Novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS One 7(2):e31235–10
Vyas VK, Berkey CD, Miyao T, Carlson M (2005) Repressors Nrg1 and Nrg2 Regulate a set of stress-responsive genes in Saccharomyces cerevisiae. Eukaryot Cell 4(11):1882–1891
Wen F-P, Guo Y-S, Hu Y et al (2016) Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 12(4):671–688
Werner-Washburne M, Braun E, Johnston GC, Singer RA (1993) Stationary-phase in the yeast Saccharomyces cerevisiae. Microbiol Rev 57(2):383–401
Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Domínguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41(11):973–981
Zhang W, Geng A (2012) Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels 5(1):1
Zhao XQ, Bai FW (2009) Mechanisms of yeast stress tolerance and its manipulation for efficient fuel ethanol production. J Biotechnol 144(1):23–30
Zhao X, Xiong L, Zhang M, Bai F (2016) Towards efficient bioethanol production from agricultural and forestry residues: Exploration of unique natural microorganisms in combination with advanced strain engineering. Bioresour Technol 215:84–91
Acknowledgements
We express our thanks to PhD. Bernardo Franco for expert assistance and technical support in the flow cytometry analysis. We also thank QFB. Miguel Roa Castañeda from Tequilera Corralejo, S.A. de C.V. for providing the Agave juice. Financial support is greatly acknowledged from the following institutions: Universidad de Guanajuato Grants: UGTO2011, UGTO 415/2014, UGTO 511/2015, UGTO 641/2015. Apoyo al Fortalecimiento de la Excelencia Académica Grants: 005/2014; 003/2015. Consejo Nacional de Ciencia y Tecnología (CONACyT): CB-2007-01. 84394; Ciencia básica: 220780, 388394; Consejo de Ciencia y Tecnología del Estado de Guanajuato (CONCyTEG): FINNOVATEC 2105, 00079. Tequilera Corralejo, S.A. de C.V., Grant: 01-2016. Naurú Idalia Vargas Maya received a scholarship from CONACyT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2016_1871_MOESM1_ESM.pdf
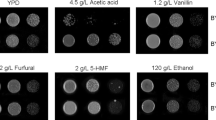
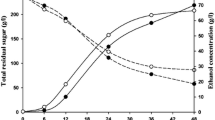
Supplementary material 1 Fig. S1 pJRC71 plasmid construction. The YNR034W-A gene was amplified by PCR using the oligonucleotides oliDYNR034W-A and oliRYNR034W-ABamHI. The HXT7 promoter region fragment was amplified from AR5 genomic DNA using oligonucleotides oliDHxt7SacI and oliRHxt7-YNR034W-A. The fusion cassette was constructed using the Double-joint PCR technique using each PCR product (HXT7 promoter region and YNR034W-A gene). The fusion PCR product was utilized in a second amplification reaction with oligonucleotides oliDHxt7SacI and oliRYNR034W-ABamHI. The final PCR fusion product (termed HXT7p-YNR034W-A) was digested with SacI and BamHI and ligated into the pJRC34 centromeric plasmid to obtain the pJRC71 plasmid. Fig. S2 AdhI C-Terminus GFP-Tag. The green fluorescent protein (GPF) and Schizosaccharomyces pombe HIS5 genes were amplified from the pFA6a-link-yEGFP-SpHIS5 plasmid (pKT128) with oligonucleotides F5GFPADH1D and R3GFPADH1R. The GFP-HIS5 cassette was used to transform the BY4741 laboratory strain. The BY4741-ADHcarboxyl-GFP strain was selected in YNB selective media. Correct cassette integration was verified by PCR using genomic DNA of each transformant and oligonucleotides ORFGFPADH1Dir and ORFGFPADH1Rev, designed outside the insertion site. Fig. S3 Ynr034w-a C-Terminus GFP-Tag. The green fluorescent protein (GPF) and Schizosaccharomyces pombe HIS5 genes were amplified from the pFA6a-link-yEGFP-SpHIS5 plasmid (pKT128) with oligonucleotides, F5GFPYNR034W-AD and R3GFPYNR034W-AR. The GFP-HIS5 cassette was used to transform the BY4741 laboratory strain. The BY4741-YNR034W-Acarboxyl-GFP strain was selected in YNB selective media. Correct cassette integration was verified by PCR using genomic DNA of each transformant and oligonucleotides ORFGFPYNR034W-AdDir and ORFGFPYNR034W-ARev, designed outside the insertion site. Fig. S4 Ynr034w A-Terminus GFP-Tag. The green fluorescent protein (GPF) and Schizosaccharomyces pombe HIS5 genes were amplified from the pFA6a-link-yEGFP-SpHIS5 plasmid (pKT128) with oligonucleotides, GFPaminoYNR034W-ADir and GFPaminoYNR034W-ARev. The GFP-HIS5 cassette was used to transform the BY4741 laboratory strain. The BY4741-YNR034W-Aamino-GFP strain was selected in YNB selective media. Correct cassette integration was verified by PCR using genomic DNA of each transformant and oligonucleotides ORFGFPYNR034W-AaminoDir and ORFGFPYNR034W-AaminoRev, designed outside the insertion site. Fig. S5 Flow cytometry analysis of BY4741-YNR034W-Aamino-GFP cells, grown in YPD supplemented with 0.6 M NaCl or 6% ethanol. Each culture was sampled at 0 h and 24 h and analyzed with a MoFlo™ XDP High-Speed Cell Sorter System (Beckman Coulter). Fluorescence was recorded from 100,000 events per sample in the FL1 channel (green fluorescence), and data analysis was performed with Summit 5.2 software. Histograms show the total fluorescence emission by cell population (FL1 channel). Representative data are from 100,000 events from biological replicates. Fig. S6 Subcellular localization of Ynr034w-a GFP. BY4741-YNR034W-Acarboxyl-GFP cells were cultured in liquid YPD for 24 h; 2x106 cells mL−1 from these cultures were inoculated in 20 mL of YPD supplemented with different stress conditions (25% glucose, 0.6 M NaCl or 6% ethanol). Cultures were grown with agitation at 28 °C. Each culture was sampled at 0 h and 24 h. Confocal microscopy images were acquired with an inverted Confocal Laser Scanning Microscope (Carl Zeiss LSM 700) with a 60X objective. Fig. S7 Cell growth of the AR5 (empty triangles), AR5-YNR034W-A (filled triangles), BY4741 (filled squares) and BY4741-YNR034 W (empty squares) yeast strains, grown in liquid YPD media. O.D. Optical Density. Data are presented as the mean of three independent experiments completed in triplicate. Fig. S8 Osmotic stress tolerance. (a) S. cerevisiae BY4741 and BY4741-YNR034W-A strains grown on YPD media plates supplemented with Hygromycin (100 µg mL−1). (b) Growth of the S. cerevisiae strains BY4741 and BY4741-YNR034W-A strains on YPD media plates supplemented with different NaCl concentrations. Fig. S9 Cell growth of the AR5 (filled symbols) and AR5-YNR034W-A (empty symbols) strains in A. tequilana Weber juice containing a sugar concentration of 180 g L−1. Data are presented as the mean of three independent experiments completed in triplicate. Cell growth data were subjected to factorial ANOVA. * Denotes a significant difference between values (p < 0.05). (PDF 4787 kb)
Rights and permissions
About this article
Cite this article
Vargas-Maya, N.I., González-Hernández, G.A., Padilla-Guerrero, I.E. et al. Overexpression of smORF YNR034W-A/EGO4 in Saccharomyces cerevisiae increases the fermentative efficiency of Agave tequilana Weber must. J Ind Microbiol Biotechnol 44, 63–74 (2017). https://doi.org/10.1007/s10295-016-1871-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1871-2