Abstract
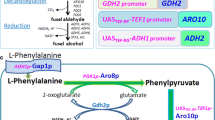
2-Phenylethanol (2-PE) is widely used in food, perfume and pharmaceutical industry, but lower production in microbes and less known regulatory mechanisms of 2-PE make further study necessary. In this study, crucial genes like ARO8 and ARO10 of Ehrlich pathway for 2-PE synthesis and key transcription factor ARO80 in Saccharomyces cerevisiae were re-regulated using constitutive promoter; in the meantime, the effect of nitrogen source in synthetic complete (SC) medium with l-phenylalanine (l-Phe) on Aro8/Aro9 and Aro10 was investigated. The results showed that aromatic aminotransferase activities of ARO8 over-expressing strains were seriously inhibited by ammonia sulfate in SC + Phe medium. Flask fermentation test demonstrated that over-expressing ARO8 or ARO10 led to about 42 % increase in 2-PE production when compared with the control strain. Furthermore, influence of transcription factors Cat8 and Mig1 on 2-PE biosynthesis was explored. CAT8 over-expression or MIG1 deletion increased in the transcription of ARO9 and ARO10. 2-PE production of CAT8 over-expressing strain was 62 % higher than that of control strain. Deletion of MIG1 also led to 2-PE biosynthesis enhancement. The strain of CAT8 over-expression and MIG1 deletion was most effective in regulating expression of ARO9 and ARO10. Analysis of mRNA levels and enzyme activities indicates that transaminase in Ehrlich pathway is the crucial target of Nitrogen Catabolize Repression (NCR). Among the engineering strains, the higher 3.73 g/L 2-PE production in CAT8 over-expressing strain without in situ product recovery suggests that the robust strain has potentiality for commercial exploitation.





Similar content being viewed by others
References
Carlson M (1999) Glucose repression in yeast. Curr Opin Microbiol 2(2):202–207
Clark GS (1990) Phenethyl alcohol. Perfum Flavor 15:37–44
Cooper TG (2002) Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev 26(3):223–238
Eshkol N, Sendovski M, Bahalul M, Katz-Ezov T, Kashi Y, Fishman A (2009) Production of 2-phenylethanol from l-phenylalanine by a stress tolerant Saccharomyces cerevisiae strain. J Appl Microbiol 106:534–542
Etschmann MMW, Bluemke W, Sell D, Schrader J (2002) Biotechnological production of 2-phenylethanol. Appl Microbiol Biotechnol 59:1–8
Etschmann MMW, Schrader J (2006) An aqueous-organic two-phase bioprocess for efficient production of the natural aroma chemicals 2-phenylethanol and 2-phenylethylacetate with yeast. Appl Microbiol Biotechnol 71(4):440–443
Fabre CE, Blanc PJ, Goma G (1998) 2-Phenylethyl alcohol: an aroma profile. Perfum Flavor 23:43–45
Fabre CE, Duviau VJ, Blanc PJ, Goma G (1995) Identification of volatile flavour compounds obtained in culture of Kluyveromyces marxianus. Biotechnol Lett 17:1207–1212
Gassenmeier K, Schieberle P (1995) Potent aromatic compounds in the crumb of wheat bread (French-type)—influence of preferments and studies on the formation of key odorants during dough processing. Z Lebensm-Unters Forsch 201:241–248
Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24(13):2519–2524
Güldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH (2002) A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res 30(6):E23–E23
Haurie V, Perrot M, Mini T, Jenö P, Sagliocco F, Boucherie H (2001) The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem 276(1):76–85
Hazelwood LA, Daran J-M, van Maris AJA, Pronk JT, Dickinson JR (2008) The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microbiol 74:2259–2266
Hedges D, Proft M, Entian KD (1995) CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol 15(4):1915–1922
Hegarty PK, Parsons R, Bamforth CW, Molzahn SW (1995) Phenyl ethanol—a factor determining lager character. In: Proc. Congr. Eur. Brew. Conv., pp 515–522
Hill JE, Meyers AM, Koerner TJ, Tzagoloff A (1993) A yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 9:163–167
Hua DL, Lin S, Li YF (2010) Enhanced 2-phenylethanol production from l-phenylalanine via in situ product adsorption. Biocatal Biotransform 28:259–266
Huang CJ, Lee SL, Chou CC (2000) Production and molar yield of 2-phenylethanol by Pichia fermentans L-5 as affected by some medium components. J Biosci Bioeng 90:142–147
Iraqui I, Vissers S, Andre B, Urrestarazu B (1999) Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae. Mol Cell Biol 19(5):3360–3371
Iraqui I, Vissers S, Cartianux M, Urrestarazu A (1998) Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet 257:238–248
Kim B, Cho BR, Hahn JS (2014) Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol Bioeng 111(1):115–124
Kradolfer P, Niederberger P, Hütter R (1982) Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases. Arch Microbiol 133:242–248
Lee K, Hahn JS (2013) Interplay of Aro80 and GATA activators in regulation of genes for catabolism of aromatic amino acids in Saccharomyces cerevisiae. Mol Microbiol 88(6):1120–1134
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Mei J, Min H, Lu Z (2009) Enhanced biotransformation of l-phenylalanine to 2-phenylethanol using an in situ product adsorption technique. Process Biochem 44(8):886–890
Pan XR, Qi HS, Mu L, Wen JP, Jia XQ (2014) Molasses wastewater for higher 2-phenylethanol production. J Agric Food Chem 62(40):9927–9935
Rahner A, Schöler A, Martens E, Gollwitzer B, Schüller H-J (1996) Dual influence of the yeast Catlp (Snflp) protein kinase on carbon source-dependent transcriptional activation of gluconeogenic genes by the regulatory gene CAT8. Nucleic Acids Res 24(12):2331–2337
Randez-Gil F, Bojunga N, Proft M, Entian KD (1997) Glucose derepression of gluconeogenic enzymes in Saccharomyces cerevisiae correlates with phosphorylation of the gene activator Cat8p. Mol Cell Biol 17(5):2502–2510
Ravasio D, Wendland J, Walther A (2013) Major contribution of the Ehrlich pathway for 2-phenylethanol/rose flavor production in Ashbya gossypii. FEMS Yeast Res 14:833–844
Romagnoli G, Knijnenburg TA, Liti G, Louis EJ, Pronk JT, Daran J-M (2014) Deletion of the Saccharomyces cerevisiae ARO8 gene, encoding an aromatic amino acid transaminase, enhances phenylethanol production from glucose. Yeast 32(1):29–45
Roth S, Kumme J, Schüller H-J (2004) Transcriptional activators Cat8 and Sip4 discriminate between sequence variants of the carbon source-responsive promoter element in the yeast Saccharomyces cerevisiae. Curr Genet 45(3):121–128
Sambrook J, Russell DW (2001) Molecular cloning, a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Schjererling P, Holmberg S (1996) Comparative amino acid sequence analysis of the C-6 zinc cluster family of transcriptional regulators. Nucleic Acids Res 24(23):4599–4607
Schüller HJ (2003) Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr Genet 43:139–160
Stahlberg A, Elbing K, Andrade-Garda JM, Sjogreen B, Forootan A, Kubista M (2008) Multiway real-time PCR gene expression profiling in yeast Saccharomyces cerevisiae reveals altered transcriptional response of ADH-genes to glucose stimuli. BMC Genom 9:170
Stark D, Kornmann H, Munch T, Sonnleitner B, Marison IW, von Stockar U (2003) Novel type of in situ extraction: use of solvent containing microcapsules for the bioconversion of 2-phenylethanol from l-phenylalanine by Saccharomyces cerevisiae. Biotechnol Bioeng 83(4):376–385
Tachibana C, Yoo JY, Tagne JB, Kacherovsky N, Lee TI, Young ET (2005) Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol 25(6):2138–2146
Teunissen AWRH, van der Berg JA, Steensma HY (1993) Physical localization of the flocculation gene FLO1 on chromosome I of Saccharomyces cerevisiae. Yeast 9(1):1–10
Urrestarazu A, Vissers S, Iraqui I, Grenson M (1998) Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet 257(2):230–237
Vaudano E, Costantini A, Cersosimo M, Del Prete V, Garcia-Moruno E (2009) Application of real-time RT-PCR to study gene expression in active dry yeast (ADY) during the rehydration phase. Int J Food Microbiol 129(1):30–36
Vuralhan Z, Morais MA, Tai S-L, Piper MDW, Pronk JT (2003) Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl Environ Microbiol 69(8):4534–4541
Walther K, Schüller H-J (2001) Adr1 and Cat8 synergistically activate the glucose-regulated alcohol dehydrogenase gene ADH2 of the yeast Saccharomyces cerevisiae. Microbiology 147(8):2037–2044
Wang H, Dong QF, Guan A, Meng C, Shi XA, Guo YH (2011) Synergistic inhibition effect of 2-phenylethanol and ethanol on bioproduction of natural 2-phenylethanol by Saccharomyces cerevisiae and process enhancement. J Biosci Bioeng 112(1):26–31
Wuster A, Babu MM (2010) Transcriptional control of the quorum sensing response in yeast. Mol Biosyst 6(1):134–141
Xu P, Hua D, Ma C (2007) Microbial transformation of propenylbenzenes for natural flavour production. Trends Biotechnol 25:571–576
Yin S, Zhou H, Xiao X, Lang T, Liang J, Wang C (2015) Improving 2-phenylethanol production via Ehrlich pathway using genetic engineered Saccharomyces cerevisiae strains. Curr Microbiol 70:762–767
Acknowledgments
This study was funded by the National Natural Science Foundation of China (Grant Number 31171754).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Z., Bai, X., Guo, X. et al. Regulation of crucial enzymes and transcription factors on 2-phenylethanol biosynthesis via Ehrlich pathway in Saccharomyces cerevisiae . J Ind Microbiol Biotechnol 44, 129–139 (2017). https://doi.org/10.1007/s10295-016-1852-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1852-5