Abstract
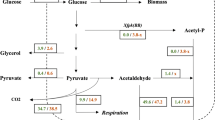
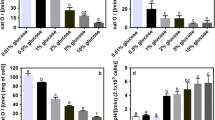
Although it is generally known that cofactors play a major role in the production of different fermentation products, their role has not been thoroughly and systematically studied. To understand the impact of cofactors on physiological functions, a systematic approach was applied, which involved redox state analysis, energy charge analysis, and metabolite analysis. Using uridine 5′-monophosphate metabolism in Saccharomyces cerevisiae as a model, we demonstrated that regulation of intracellular the ratio of NADPH to NADP+ not only redistributed the carbon flux between the glycolytic and pentose phosphate pathways, but also regulated the redox state of NAD(H), resulting in a significant change of ATP, and a significantly altered spectrum of metabolic products.







Similar content being viewed by others
References
Bailey JE (1991) Toward a science of metabolic engineering. Science 252:1668–1675
Bakker BM, Overkamp KM, van Maris AJ et al (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37
Berrios-Rivera SJ, Bennett GN, San KY (2003) The effect of carbon sources and lactate dehydrogenase deletion on 1,2-propanediol production in Escherichia coli. Metab Eng 4:217–229
Berrios-Rivera SJ, Bennett GN, San KY (2003) The effect of increasing NADH availability on the redistribution of metabolic fluxes in Escherichia coli chemostat cultures. Metab Eng 4:230–237
Berrios-Rivera SJ, San KY, Bennett GN (2003) The effect of NAPRTase overexpression on the total levels of NAD+, the NADH-NAD+ ratio, and the distribution of metabolites in Escherichia coli. Metab Eng 4:238–247
Chen Y, Li SY, Xiong J et al (2010) The mechanisms of citrate on regulating the distribution of carbon flux in the biosynthesis of uridine 5′-monophosphate by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 86:75–81
DeLuna A, Avendano A, Riego L et al (2001) NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. Purification, kinetic properties, and physiological roles. J Biol Chem 276:43775–43783
Du CY, Yan H, Zhang YP et al (2006) Use oxidoreduction potentials an indicator to regulate 1,3-propanediol fermentation by Klebsiella pneumoniae. Appl Microbiol Biotechnol 69:554–563
Dos Santos MM, Raghevendran V, Kotter P et al (2004) Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab Eng 6:352–363
Garrigues C, Loubiere P, Lindley ND et al (1997) Control of the shift from homolactic acid to mixed-acid fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J Bacteriol 179:5282–5287
Heux S, Cachon R, Dequin S (2006) Cofactor engineering in Saccharomyces cerevisiae: expression of a H(2)O-forming NADH oxidase and impact on redox metabolism. Metab Eng 8:303–314
Jol SJ, Kümmel A, Terzer M et al (2012) System-level insights into yeast metabolism by thermodynamic analysis of elementary flux modes. PLoS Comput Biol. doi:10.1371/journal.pcbi.1002415
Kamada N, Yasuhara A, Ikeda M (2003) Significance of the non-oxidative route of the pentose phosphate pathway for supplying carbon to the purine-nucleotide pathway in Corynebacterium ammoniagenes. J Ind Microbiol Biotechnol 30:129–132
Larsson C, Nilsson A, Blomberg A et al (1997) Glycolytic flux is conditionally correlated with ATP concentration in Saccharomyces cerevisiae: a chemostat study under carbon- or nitrogen-limiting conditions. J Bacteriol 179:7243–7250
Liu W, Wang P (2007) Cofactor regeneration for sustainable enzymatic biosynthesis. Biotechnol Adv 25:369–384
Poulsen R, Nohr J, Douthwaite S et al (2005) Increased NADPH concentration obtained by metabolic engineering of the pentose phosphate pathway in Aspergillus niger. FEBS J 72:1313–1325
Remize F, Barnavon L, Dequin S (2001) Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatases, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab Eng 3:301–312
Sánchez AM, Bennett GN, San KY (2005) Effect of different levels of NADH availability on metabolic fluxes of Escherichia coli chemostat cultures in defined medium. J Biotechnol 117:395–405
Stephanopoulos GN, Vallino JJ (1991) Network rigidity and metabolic engineering in metabolite overproduction. Science 252:1675–1681
Vaseghi S, Baumeister A, Rizzi M, Reuss M (1999) In vivo dynamics of the pentose phosphate pathway in Saccharomyces cerevisiae. Metab Eng 1:128–140
Verho R, Londesborough J, Penttilä M et al (2003) Engineering redox cofactor regeneration for improved pentose fermentation in Saccharomyces cerevisiae. Appl Environ Microbiol 10:5892–5897
Wang X, Wang XW, Yin MX et al (2007) Production of uridine 5′-monophosphate by Corynebacterium ammoniagenes ATCC 6872 using a statistically improved biocatalytic process. Appl Microbiol Biotechnol 76:321–328
Zhang YP, Huang ZH, Dua CH et al (2009) Introduction of an NADH regeneration system into Klebsiella oxytoca leads to an enhanced oxidative and reductive metabolism of glycerol. Metab Eng 11:101–106
Zhang Z, Yu J, Stanton RC (2000) A method for determination of pyridine nucleotides using a single extract. Anal Biochem 285:163–167
Zhou JW, Liu LM, Shi ZP et al (2009) ATP in current biotechnology: Regulation, applications and perspectives. Biotechnol Adv 27:94–101
Acknowledgments
This work was supported by the National Outstanding Youth Foundation of China (Grant No. 21025625), the National High-Tech Research and Development Program of China (863) (Grant No. 2012AA021200), the National Basic Research Program of China (973) (Grant No. 2011CBA00806), the National Key Technology R&D Program (2012BAI44G01), the Program of Changjiang Scholars and Innovative in University (Grant No. IRT1066), the National Natural Science Foundation of China, the Youth Program (Grant No. 21106070), the Jiangsu Provincial Natural Science Foundation of China (Grant No. SBK 201150207), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the State Key Laboratory of Motor Vehicle Biofuel Technology (Grant No. 2013003)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, Y., Liu, Q., Chen, X. et al. Redirecting metabolic flux in Saccharomyces cerevisiae through regulation of cofactors in UMP production. J Ind Microbiol Biotechnol 42, 577–583 (2015). https://doi.org/10.1007/s10295-014-1536-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-014-1536-y