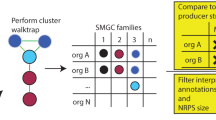
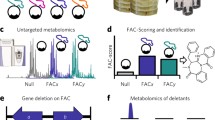
Abstract
Fungi are prolific producers of secondary metabolites (SMs) that show a variety of biological activities. Recent advances in genome sequencing have shown that fungal genomes harbor far more SM gene clusters than are expressed under conventional laboratory conditions. Activation of these “silent” gene clusters is a major challenge, and many approaches have been taken to attempt to activate them and, thus, unlock the vast treasure chest of fungal SMs. This review will cover recent advances in genome mining of SMs in Aspergillus nidulans. We will also discuss current updates in gene annotation of A. nidulans and recent developments in A. nidulans as a molecular genetic system, both of which are essential for rapid and efficient experimental verification of SM gene clusters on a genome-wide scale. Finally, we will describe advances in the use of A. nidulans as a heterologous expression system to aid in the analysis of SM gene clusters from other fungal species that do not have an established molecular genetic system.

Similar content being viewed by others
References
Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CCC (2012) Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc 134(19):8212–8221. doi:10.1021/ja3016395
Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, Blicher LH, Gotfredsen CH, Larsen TO, Nielsen KF, Mortensen UH (2013) Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci USA 110(1):E99–E107. doi:10.1073/pnas.1205532110
Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, Crabtree J, Howarth C, Orvis J, Shah P, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G, Wortman JR (2012) The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res 40 (D1):D653–D659. doi:10.1093/nar/gkr875
Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G (2010) The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res 38:D420–D427. doi:10.1093/nar/gkp751
Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C (2007) Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol 3(4):213–217. doi:10.1038/nchembio869
Bode HB, Bethe B, Hofs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3(7):619–627. doi:10.1002/1439-7633(20020703)3:7<619:aid-cbic619>3.0.co;2-9
Bok JW, Chiang YM, Szewczyk E, Reyes-Domingez Y, Davidson AD, Sanchez JF, Lo H-C, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP (2009) Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol 5(7):462–464. doi:10.1038/nchembio.177
Bok JW, Hoffmeister D, Maggio-Hall LA, Murillo R, Glasner JD, Keller NP (2006) Genomic mining for Aspergillus natural products. Chem Biol 13(1):31–37. doi:10.1016/j.chembiol.2005.10.008
Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D (2007) Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet Biol 44(11):1134–1145. doi:10.1016/j.fgb.2006.12.010
Brakhage AA (2013) Regulation of fungal secondary metabolism. Nat Rev Microbiol 11(1):21–32. doi:10.1038/nrmicro2916
Brakhage AA, Schuemann J, Bergmann S, Scherlach K, Schroeckh V, Hertweck C (2008) Activation of fungal silent gene clusters: a new avenue to drug discovery. Prog Drug Res 66:12
Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ (1996) Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA 93(4):1418–1422. doi:10.1073/pnas.93.4.1418
Brown NA, de Gouvea PF, Krohn NG, Savoldi M, Goldman GH (2013) Functional characterisation of the non-essential protein kinases and phosphatases regulating Aspergillus nidulans hydrolytic enzyme production. Biotechnol Biofuels 6. doi:10.1186/1754-6834-6-91
Chiang YM, Chang SL, Oakley BR, Wang CCC (2011) Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol 15(1):137–143. doi:10.1016/j.cbpa.2010.10.011
Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, Sung CT, Wang CCC, Oakley BR (2013) An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc 135(20):7720–7731. doi:10.1021/ja401945a
Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CCC, Oakley BR (2010) Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol 76(7):2067–2074. doi:10.1128/aem.02187-09
Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC (2009) A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone in Aspergillus nidulans. J Am Chem Soc 131(8):2965–2970. doi:10.1021/ja8088185
Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC (2008) Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol 15(6):527–532. doi:10.1016/j.chembiol.2008.05.010
De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, Osmani SA (2013) Functional analysis of the Aspergillus nidulans kinome. PLoS ONE 8(3)
Eisendle M, Oberegger H, Zadra I, Haas H (2003) The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N-5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol Microbiol 49(2):359–375. doi:10.1046/j.1365-2958.2003.03586.x
Fujii I, Watanabe A, Sankawa U, Ebizuka Y (2001) Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol 8(2):189–197. doi:10.1016/s1074-5521(00)90068-1
Fujii I, Yoshida N, Shimomaki S, Oikawa H, Ebizuka Y (2005) An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem Biol 12(12):1301–1309. doi:10.1016/j.chembiol.2005.09.015
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D’Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Penalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A-fumigatus and A-oryzae. Nature 438(7071):1105–1115. doi:10.1038/nature04341
Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24(2):393–416. doi:10.1039/b603084j
Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G (2013) Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol 13. doi:10.1186/1471-2180-13-91
Kasahara K, Fujii I, Oikawa H, Ebizuka Y (2006) Expression of Alternaria solani PKSF generates a set of complex reduced-type polyketides with different carbon-lengths and cyclization. ChemBioChem 7(6):920–924. doi:10.1002/cbic.200600034
Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND (2010) SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47(9):736–741. doi:10.1016/j.fgb.2010.06.003
Klejnstrup ML, Frandsen RJN, Holm DK, Nielsen MT, Mortensen UH, Larsen TO, Nielsen JB (2012) Genetics of polyketide metabolism in Aspergillus nidulans. Metabolites 2(1):100–133. doi:10.3390/metabo2010100
Li Y, Chooi Y-H, Sheng Y, Valentine JS, Tang Y (2011) Comparative characterization of fungal anthracenone and naphthacenedione biosynthetic pathways reveals an alpha-hydroxylation-dependent Claisen-like cyclization catalyzed by a dimanganese thioesterase. J Am Chem Soc 133(39):15773–15785. doi:10.1021/ja206906d
Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CCC (2012) Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc 134(10):4709–4720. doi:10.1021/ja209809t
Maccabe AP, Vanliempt H, Palissa H, Unkles SE, Riach MBR, Pfeifer E, Vondohren H, Kinghorn JR (1991) Delta-(l-alpha-aminoadipyl)-l-cysteinyl-d-valine synthetase from Aspergillus nidulans: molecular characterization of the Acva gene encoding the 1st enzyme of the penicillin biosynthetic-pathway. J Biol Chem 266(19):12646–12654
Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto KI, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu JJ, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438(7071):1157–1161. doi:10.1038/nature04300
Maiya S, Grundmann A, Li SM, Turner G (2006) The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem 7(7):1062–1069. doi:10.1002/cbic.200600003
Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi:10.1093/nar/gkr466
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75(3):311–335. doi:10.1021/np200906s
Nielsen ML, Nielsen JB, Rank C, Klejnstrup ML, Holm DK, Brogaard KH, Hansen BG, Frisvad JC, Larsen TO, Mortensen UH (2011) A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol Lett 321(2):157–166. doi:10.1111/j.1574-6968.2011.02327.x
Nielsen MT, Nielsen JB, Anyaogu DC, Holm DK, Nielsen KF, Larsen TO, Nortensen UH (2013) Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR-based approach. PLoS ONE 8(8):e72871. doi:10.1371/journal.pone.0072871
Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafton A, Latge JP, Li WX, Lord A, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O’Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, de Cordoba SR, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, de Aldana CRV, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW (2005) Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438(7071):1151–1156. doi:10.1038/nature04332
Nitsche BM, Crabtree J, Cerqueira GC, Meyer V, Ram AFJ, Wortman JR (2011) New resources for functional analysis of omics data for the genus Aspergillus. BMC Genomics 12. doi:10.1186/1471-2164-12-486
Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, Andersen MR, Bendtsen JD, Benen JAE, van den Berg M, Breestraat S, Caddick MX, Contreras R, Cornell M, Coutinho PM, Danchin EGJ, Debets AJM, Dekker P, van Dijck PWM, van Dijk A, Dijkhuizen L, Driessen AJM, d’Enfert C, Geysens S, Goosen C, Groot GSP, de Groot PWJ, Guillemette T, Henrissat B, Herweijer M, van den Hombergh J, van den Hondel C, van der Heijden R, van der Kaaij RM, Klis FM, Kools HJ, Kubicek CP, van Kuyk PA, Lauber J, Lu X, van der Maarel M, Meulenberg R, Menke H, Mortimer MA, Nielsen J, Oliver SG, Olsthoorn M, Pal K, van Peij N, Ram AFJ, Rinas U, Roubos JA, Sagt CMJ, Schmoll M, Sun JB, Ussery D, Varga J, Vervecken W, de Vondervoort P, Wedler H, Wosten HAB, Zeng AP, van Ooyen AJJ, Visser J, Stam H (2007) Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol 25(2):221–231. doi:10.1038/nbt1282
Rohlfs M, Albert M, Keller NP, Kempken F (2007) Secondary chemicals protect mould from fungivory. Biol Lett 3(5):523–525. doi:10.1098/rsbl.2007.0338
Sanchez JF, Chiang Y-M, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC (2010) Molecular genetic analysis of the orsellinic acid/F9775 gene cluster of Aspergillus nidulans. Mol BioSyst 6(3):587–593. doi:10.1039/b904541d
Sanchez JF, Entwistle R, Corcoran D, Oakley BR, Wang CCC (2012) Identification and molecular genetic analysis of the cichorine gene cluster in Aspergillus nidulans. MedChemComm 3(8):997–1002. doi:10.1039/c2md20055d
Sanchez JF, Entwistle R, Hung J-H, Yaegashi J, Jain S, Chiang Y-M, Wang CCC, Oakley BR (2011) Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc 133(11):4010–4017. doi:10.1021/ja1096682
Sanchez JF, Somoza AD, Keller NP, Wang CCC (2012) Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat Prod Rep 29(3):351–371. doi:10.1039/c2np00084a
Scherlach K, Sarkar A, Schroeckh V, Dahse H-M, Roth M, Brakhage AA, Horn U, Hertweck C (2011) Two induced fungal polyketide pathways converge into antiproliferative spiroanthrones. ChemBioChem 12(12):1836–1839. doi:10.1002/cbic.201100132
Scherlach K, Schuemann J, Dahse HM, Hertweck C (2010) Aspernidine A and B, prenylated isoindolinone alkaloids from the model fungus Aspergillus nidulans. J Antibiot 63(7):375–377. doi:10.1038/ja.2010.46
Schroeckh V, Scherlach K, Nuetzmann H-W, Shelest E, Schmidt-Heck W, Schuemann J, Martin K, Hertweck C, Brakhage AA (2009) Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci USA 106(34):14558–14563. doi:10.1073/pnas.0901870106
Somoza AD, Lee KH, Chiang YM, Oakley BR, Wang CCC (2012) Reengineering an azaphilone biosynthesis pathway in Aspergillus nidulans to create lipoxygenase inhibitors. Org Lett 14(4):972–975. doi:10.1021/ol203094k
Son S, Osmani SA (2009) Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, Fcp1. Eukaryot Cell 8(4):573–585. doi:10.1128/ec.00346-08
Soukup AA, Chiang Y-M, Bok JW, Reyes-Dominguez Y, Oakley BR, Wang CCC, Strauss J, Keller NP (2012) Overexpression of the Aspergillus nidulans histone 4 acetyltransferase EsaA increases activation of secondary metabolite production. Mol Microbiol 86(2):314–330. doi:10.1111/j.1365-2958.2012.08195.x
Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CCC, Oakley BR (2008) Identification and characterization of the asperthecin gene cluster of Aspergillus nidulans. Appl Environ Microbiol 74(24):7607–7612. doi:10.1128/aem.01743-08
Tsunematsu Y, Ki Ishiuchi, Hotta K, Watanabe K (2013) Yeast-based genome mining, production and mechanistic studies of the biosynthesis of fungal polyketide and peptide natural products. Nat Prod Rep 30(8):1139–1149. doi:10.1039/c3np70037b
Wang CCC, Chiang Y-M, Kuo P-L, Chang J-K, Hsu Y-L (2008) Norsolorinic acid from Aspergillus nidulans inhibits the proliferation of human breast adenocarcinoma MCF-7 cells via fas-mediated pathway. Basic Clin Pharmacol Toxicol 102(6):491–497. doi:10.1111/j.1742-7843.2008.00237.x
Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y (1999) Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Lett 40(1):91–94. doi:10.1016/s0040-4039(98)80027-0
Wiemann P, Keller NP (2013) Strategies for mining fungal natural products. J Ind Microbiol Biotechnol (this issue). doi:10.1007/s10295-013-1366-3
Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, von Dohren H, Doonan J, Driessen AJM, Durek P, Espeso E, Fekete E, Flipphi M, Estrada CG, Geysens S, Goldman G, de Groot PWJ, Hansen K, Harris SD, Heinekamp T, Helmstaedt K, Henrissat B, Hofmann G, Homan T, Horio T, Horiuchi H, James S, Jones M, Karaffa L, Karanyi Z, Kato M, Keller N, Kelly DE, Kiel J, Kim JM, van der Klei IJ, Klis FM, Kovalchuk A, Krasevec N, Kubicek CP, Liu B, MacCabe A, Meyer V, Mirabito P, Miskei M, Mos M, Mullins J, Nelson DR, Nielsen J, Oakley BR, Osmani SA, Pakula T, Paszewski A, Paulsen I, Pilsyk S, Pocsi I, Punt PJ, Ram AFJ, Ren QH, Robellet X, Robson G, Seiboth B, van Solingen P, Specht T, Sun JB, Taheri-Talesh N, Takeshita N, Ussery D, Vankuyk PA, Visser H, de Vondervoort P, de Vries RP, Walton J, Xiang X, Xiong Y, Zeng AP, Brandt BW, Cornell MJ, van den Hondel C, Visser J, Oliver SG, Turner G (2009) The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet Biol 46:S2–S13. doi:10.1016/j.fgb.2008.12.003
Yaegashi J, Praseuth MB, Tyan S-W, Sanchez JF, Entwistle R, Chiang Y-M, Oakley BR, Wang CCC (2013) Molecular genetic characterization of the biosynthesis cluster of a prenylated isoindolinone alkaloid aspernidine A in Aspergillus nidulans. Org Lett 15(11):2862–2865. doi:10.1021/ol401187b
Yeh HH, Chiang YM, Entwistle R, Ahuja M, Lee KH, Bruno KS, Wu TK, Oakley BR, Wang CCC (2012) Molecular genetic analysis reveals that a nonribosomal peptide synthetase-like (NRPS-like) gene in Aspergillus nidulans is responsible for microperfuranone biosynthesis. Appl Microbiol Biotechnol 96(3):739–748. doi:10.1007/s00253-012-4098-9
Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y (2013) Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth Biol. doi:10.1021/sb400048b
Yin WB, Keller NP (2011) Transcriptional regulatory elements in fungal secondary metabolism. J Microbiol 49(3):329–339. doi:10.1007/s12275-011-1009-1
Yu JH, Leonard TJ (1995) Sterigmatocystin biosynthesis in Aspergillus nidulans requires a novel type I polyketide synthase. J Bacteriol 177(16):4792–4800
Acknowledgments
C.C.C.W and B.R.O gratefully acknowledge the National Institutes of Health (GM084077) for supporting research on Aspergillus nidulans secondary metabolism. We also thank Elizabeth Oakley and Dr. James Sanchez for their editorial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yaegashi, J., Oakley, B.R. & Wang, C.C.C. Recent advances in genome mining of secondary metabolite biosynthetic gene clusters and the development of heterologous expression systems in Aspergillus nidulans . J Ind Microbiol Biotechnol 41, 433–442 (2014). https://doi.org/10.1007/s10295-013-1386-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1386-z