Abstract
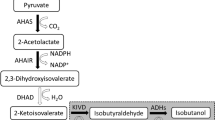
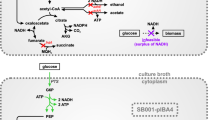
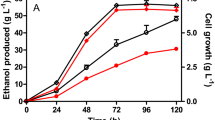
A homobutanol fermentation pathway was engineered in a derivative of Escherichia coli B (glucose [glycolysis] => 2 pyruvate + 2 NADH; pyruvate [pyruvate dehydrogenase] => acetyl-CoA + NADH; 2 acetyl-CoA [butanol pathway enzymes] + 4 NADH => butanol; summary stoichiometry: glucose => butanol). Initially, the native fermentation pathways were eliminated from E. coli B by deleting the genes encoding for lactate dehydrogenase (ldhA), acetate kinase (ackA), fumarate reductase (frdABCD), pyruvate formate lyase (pflB), and alcohol dehydrogenase (adhE), and the pyruvate dehydrogenase complex (aceEF-lpd) was anaerobically expressed through promoter replacement. The resulting strain, E. coli EG03 (ΔfrdABCD ΔldhA ΔackA ΔpflB Δ adhE ΔpdhR ::pflBp6-aceEF-lpd ΔmgsA), could generate 4 NADH for every glucose oxidized to two acetyl-CoA through glycolysis and the pyruvate dehydrogenase complex. However, EG03 lost its ability for anaerobic growth due to the lack of NADH oxidation pathways. When the butanol pathway genes that encode for acetyl-CoA acetyltransferase (thiL), 3-hydroxybutyryl-CoA dehydrogenase (hbd), crotonase (crt), butyryl-CoA dehydrogenase (bcd, etfA, etfB), and butyraldehyde dehydrogenase (adheII) were cloned from Clostridium acetobutylicum ATCC 824, and expressed in E. coli EG03, a balanced NADH oxidation pathway was established for homobutanol fermentation (glucose => 4 NADH + 2 acetyl-CoA => butanol). This strain was able to convert glucose to butanol (1,254 mg l−1) under anaerobic condition.



Similar content being viewed by others
References
Atsumi S, Cann AF, Connor MR, Shen CR, Smith KM, Brynildsen MP, Chou KJY, Hanai T, Liao JC (2008) Metabolic engineering of Escherichia coli for 1-butanol production. Metab Eng 10:305–311
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) (1987) Current protocols in molecular biology, vol 1. Greene Publishing Associates/Wiley Interscience, New York
Causey TB, Zhou S, Shanmugam KT, Ingram LO (2003) Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homo-acetate production. Proc Natl Acad Sci USA 100:825–832
Chen K, Iverson AG, Garza EA, Grayburn WS, Zhou S (2010) Metabolic evolution of non-transgenic Escherichia coli SZ420 for enhanced homoethanol fermentation from xylose. Biotechnol Lett 32:87–96
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Rev 63:223–234
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645
Energy Information Administration (2007) Biofuels in the U.S. transportation sector. 22 Mar 2011. http://www.eia.doe.gov/oiaf/analysispaper/biomass.html
Garza E, Finan C, Iverson A, Zhou S (2011) Extension temperature of 60 °C required for PCR of large DNA fragments (>5 kb) from a low GC bacterium Clostridium acetobutylicum. World J Microbiol Biotechnol 27:449–451
Inui M, Suda M, Kimura S, Yasuda K, Suzuki H, Toda H, Yamamoto S, Okino S, Suzuki N, Yukawa H (2008) Expression of Clostridium acetobutylicum butanol synthetic genes in Escherichia coli. Appl Microbiol Biotechnol 77:1305–1316
Jang YS, Lee J, Malaviya A, Seung DY, Cho JH, Lee SY (2012) Butanol production from renewable biomass: rediscovery of metabolic pathways and metabolic engineering. J Biotechnol 7:186–198
Jiang Y, Xu C, Dong F, Yang Y, Jiang W, Yang S (2009) Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab Eng 11:284–291
Jones DT, Woods DR (1986) Acetone-butanol fermentation revisited. Microbiol Rev 50(4):484–524
Kataoka N, Tajima T, Kato J, Rachadech W, Vangnai AS (2011) Development of butanol-tolerant Bacillus subtilis strain GRSW2-B1 as a potential bioproduction host. AMB Express 1:10
Lee J, Jang Y, Choi SJ, Im JA, Song H, Cho JH, Seung DY, Papoutsakis ET, Bennett GN, Lee S (2012) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation. Appl Environ Microbiol 78(5):1416–1423
Lee JY, Yang KS, Jang SA, Sung BH, Kim SC (2011) Engineering butanol-tolerance in Escherichia coli with artificial transcription factor libraries. Biotechnol Bioeng 108:742–749
Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS (2008) Fermentative butanol production by Clostridia. Biotechnol Bioeng 101(2):209–228
Manow R, Wang J, Wang Y, Zhao J, Garza E, Iverson A, Finan C, Grayburn S, Zhou S (2012) Partial deletion of rng (RNase G)-enhanced homoethanol fermentation of xylose by the non-transgenic Escherichia coli RM10. J Ind Microbiol Biotechnol. doi:10.1007/s10295-012-1100-6
Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Press, Cold Spring Harbor, New York
Pospai G, Koob MD, Kirkpatrick HA, Blattner FC (1997) Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol 179:4426–4428
Qurenshi N, Blaschek HP (2001) ABE production from corn: a recent economic evaluation. J Ind Microbiol Biotechnol 27:292–297
Ruhl J, Schmid A, Blank LM (2009) Selected Pseudomonas putida strains able to grow in the presence of high butanol concentrations. Appl Environ Microbiol 75(13):4653–4656
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Press, Cold Spring Harbor, New York
Shen CR, Lan EI, Dekishima Y, Baez A, Cho K, Liao JC (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol 77(9):2905–2915
Shapovalov OI, Ashkinazi LA (2008) Biobutanol: biofuel of second generation. Russian J Appl Chem 81(12):2232–2236
Sillers R, Al-Hinai MA, Papoutsakis ET (2009) Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectively in Clostridium acetobutylicum fermentations. Biotechnol Bioeng 102:38–49
Steen EJ, Chan R, Prasad N, Myers S, Petzold CJ, Redding A, Ouellet M, Keasling JD (2008) Metabolic engineering of Saccharomyces cerevisiae for production of n-butanol. Microb Cell Fact 7:36
Vasconcelos I, Girbal L, Soucaille P (1994) Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures or glucose and glycerol. J Bacteriol 176(5):1443–1450
Wang Y, Manow R, Finan C, Wang J, Garza E, Zhou S (2011) Adaptive evolution of non-transgenic Escherichia coli KC01 for improved ethanol tolerance and homoethanol fermentation from xylose. J Ind Microbiol Biotechnol 38:1371–1377
Zhou S, Iverson AG, Grayburn WS (2008) Engineering a native homoethanol pathway in Escherichia coli B for ethanol production. Biotechnol Lett 30(2):335–342
Zhou S, Iverson AG, Grayburn WS (2010) Doubling the catabolic reducing power (NADH) output of Escherichia coli fermentation for production of reduced products. Biotechnol Progress 26(1):45–51
Acknowledgments
This research was supported by a summer artistry and research grant, biological research incentive fund of Northern Illinois University, a grant from the China National Natural Science Foundation (NSFC31070094), a grant from the Hubei Provincial Natural Science Foundation (2011CDA008) and the Chutian Scholar Program, P.R. China.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Garza, E., Zhao, J., Wang, Y. et al. Engineering a homobutanol fermentation pathway in Escherichia coli EG03. J Ind Microbiol Biotechnol 39, 1101–1107 (2012). https://doi.org/10.1007/s10295-012-1151-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-012-1151-8