Abstract
Two genes that encode α-amylases from two Anoxybacillus species were cloned and expressed in Escherichia coli. The genes are 1,518 bp long and encode 506 amino acids. Both sequences are 98% similar but are distinct from other well-known α-amylases. Both of the recombinant enzymes, ASKA and ADTA, were purified using an α-CD–Sepharose column. They exhibited an optimum activity at 60°C and pH 8. Both amylases were stable at pH 6–10. At 60°C in the absence of Ca2+, negligible reduction in activity for up to 48 h was observed. The activity half-life at 65°C was 48 and 3 h for ASKA and ADTA, respectively. In the presence of Ca2+ ions, both amylases were highly stable for at least 48 h and had less than a 10% decrease in activity at 70°C. Both enzymes exhibited similar end-product profiles, and the predominant yield was maltose (69%) from starch hydrolysis. To the best of our knowledge, most α-amylases that produce high levels of maltose are active at an acidic to neutral pH. This is the first report of two thermostable, alkalitolerant recombinant α-amylases from Anoxybacillus that produce high levels of maltose and have an atypical protein sequence compared with known α-amylases.





Similar content being viewed by others
References
Arikan B (2008) Highly thermostable, thermophilic, alkaline, SDS and chelator resistant amylase from a thermophilic Bacillus sp. isolate A3–15. Bioresour Technol 99:3071–3076. doi:10.1016/j.biortech.2007.06.019
Asoodeh A, Chamani J, Lagzian M (2010) A novel thermostable, acidophilic α-amylase from a new thermophilic “Bacillus sp. Ferdowsicous” isolated from Ferdows hot mineral spring in Iran: purification and biochemical characterization. Int J Biol Macromol 46:289–297. doi:10.1016/j.ijbiomac.2010.01.013
Ballschmiter M, Armbrecht M, Ivanova K, Antranikian G, Liebl W (2005) AmyA, an α-amylase with β-cyclodextrin-forming activity, and AmyB from the thermoalkaliphilic organism Anaerobranca gottschalkii: two α-amylases adapted to their different cellular localizations. Appl Environ Microbiol 71:3709–3715. doi:10.1128/aem.71.7.3709-3715.2005
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: Signal P 3.0. J Mol Biol 340:783–795. doi:10.1016/j.jmb.2004.05.028
Bolton DJ, Kelly CT, Fogarty WM (1997) Purification and characterization of the α-amylase of Bacillus flavothermus. Enzyme Microb Technol 20:340–343. doi:10.1016/S0141-0229(96)00147-0
Busby S, Ebright RH (1994) Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743–746. doi:10.1016/0092-8674(94)90063-9
Chai YY, Kahar UM, Salleh MM, Illias RM, Goh KM (2011) Isolation and characterisation of pullulan-degrading Anoxybacillus species isolated from Malaysian hot springs. Environ Technol. doi:10.1080/09593330.2011.618935
Collins BS, Kelly CT, Fogarty WM, Doyle EM (1993) The high maltose-producing α-amylase of the thermophilic actinomycete, Thermomonospora curvata. Appl Microbiol Biotechnol 39:31–35. doi:10.1007/bf00166844
Declerck N, Machius M, Chambert R, Wiegand G, Huber R, Gaillardin C (1997) Hyperthermostable mutants of Bacillus licheniformis alpha-amylase: thermodynamic studies and structural interpretation. Protein Eng 10:541–549. doi:10.1093/protein/10.5.541
Declerck N, Machius M, Joyet P, Wiegand G, Huber R, Gaillardin C (2003) Hyperthermostabilization of Bacillus licheniformis α-amylase and modulation of its stability over a 50°C temperature range. Protein Eng 16:287–293. doi:10.1093/proeng/gzg032
Declerck N, Machius M, Wiegand G, Huber R, Gaillardin C (2000) Probing structural determinants specifying high thermostability in Bacillus licheniformis α-amylase. J Mol Biol 301:1041–1057. doi:10.1006/jmbi.2000.4025
Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B (2003) Microbial α-amylases: a biotechnological perspective. Process Biochem 38:1599–1616. doi:10.1016/S0032-9592(03)00053-0
Hagihara H, Igarashi K, Hayashi Y, Endo K, Ikawa-Kitayama K, Ozaki K, Kawai S, Ito S (2001) Novel α-amylase that is highly resistant to chelating reagents and chemical oxidants from the alkaliphilic Bacillus isolate KSM-K38. Appl Environ Microbiol 67:1744–1750. doi:10.1128/aem.67.4.1744-1750.2001
Hashim SO, Delgado OD, Martínez MA, Kaul RH, Mulaa FJ, Mattiasson B (2005) Alkaline active maltohexaose-forming α-amylase from Bacillus halodurans LBK 34. Enzyme Microb Technol 36:139–146. doi:10.1016/j.enzmictec.2004.07.017
Hmidet N, Bayoudh A, Berrin JG, Kanoun S, Juge N, Nasri M (2008) Purification and biochemical characterization of a novel α-amylase from Bacillus licheniformis NH1: cloning, nucleotide sequence and expression of amyN gene in Escherichia coli. Process Biochem 43:499–510. doi:10.1016/j.procbio.2008.01.017
Janecek S (2002) How many conserved sequence regions are there in the α-amylase family? Biologia 57:29–41
Kiran KK, Chandra TS (2008) Production of surfactant and detergent-stable, halophilic, and alkalitolerant alpha-amylase by a moderately halophilic Bacillus sp strain TSCVKK. Appl Microbiol Biotechnol 77:1023–1031. doi:10.1007/s00253-007-1250-z
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Li S, Zuo Z, Niu D, Singh S, Permaul K, Prior BA, Shi G, Wang Z (2011) Gene cloning, heterologous expression, and characterization of a high maltose-producing α-amylase of Rhizopus oryzae. Appl Biochem Biotechnol 164:581–592. doi:10.1007/s12010-011-9159-5
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi:10.1021/ac60147a030
Murakami S, Nagasaki K, Nishimoto H, Shigematu R, Umesaki J, Takenaka S, Kaulpiboon J, Prousoontorn M, Limpaseni T, Pongsawasdi P, Aoki K (2008) Purification and characterization of five alkaline, thermotolerant, and maltotetraose-producing α-amylases from Bacillus halodurans MS-2–5, and production of recombinant enzymes in Escherichia coli. Enzyme Microb Technol 43:321–328. doi:10.1016/j.enzmictec.2008.05.006
Palva I, Pettersson RF, Kalkkinen N, Lehtovaara P, Sarvas M, Soderlund H, Takkinen K, Kaariainen L (1981) Nucleotide sequence of the promoter and NH2-terminal signal peptide region of the α-amylase gene from Bacillus amyloliquefaciens. Gene 15:43–51. doi:10.1016/0378-1119(81)90103-7
Poli A, Salerno A, Laezza G, di Donato P, Dumontet S, Nicolaus B (2009) Heavy metal resistance of some thermophiles: potential use of α-amylase from Anoxybacillus amylolyticus as a microbial enzymatic bioassay. Res Microbiol 160:99–106. doi:10.1016/j.resmic.2008.10.012
Prakash B, Vidyasagar M, Madhukumar MS, Muralikrishna G, Sreeramulu K (2009) Production, purification, and characterization of two extremely halotolerant, thermostable, and alkali-stable α-amylases from Chromohalobacter sp. TVSP 101. Process Biochem 44:210–215. doi:10.1016/j.procbio.2008.10.013
Saito N (1973) A thermophilic extracellular α-amylase from Bacillus licheniformis. Arch Biochem Biophys 155:290–298
Shafiei M, Ziaee AA, Amoozegar MA (2011) Purification and characterization of an organic-solvent-tolerant halophilic α-amylase from the moderately halophilic Nesterenkonia sp. strain F. J Ind Microbiol Biotechnol 38:275–281. doi:10.1007/s10295-010-0770-1
Sharma A, Satyanarayana T (2010) High maltose-forming, Ca2+-independent and acid stable α-amylase from a novel acidophilic bacterium, Bacillus acidicola. Biotechnol Lett 32:1503–1507. doi:10.1007/s10529-010-0322-9
Sivaramakrishnan S, Gangadharan D, Nampoothiri KM, Soccol CR, Pandey A (2006) α-Amylases from microbial sources—an overview on recent developments. Food Technol Biotechnol 44:173–184
Stam MR, Danchin EGJ, Rancurel C, Coutinho PM, Henrissat B (2006) Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Eng Des Sel 19:555–562. doi:10.1093/protein/gzl044
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Tanaka A, Hoshino E (2003) Secondary calcium-binding parameter of Bacillus amyloliquefaciens α-amylase obtained from inhibition kinetics. Biosci Bioeng 96:262–267. doi:10.1016/S1389-1723(03)80191-3
Tao X, Jang MS, Kim KS, Yu Z, Lee YC (2008) Molecular cloning, expression and characterization of alpha-amylase gene from a marine bacterium Pseudoalteromonas sp. MY-1. Indian J Biochem Biophys 45:305–309
van der Maarel MJEC, van der Veen B, Uitdehaag JCM, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol 94:137–155. doi:10.1016/S0168-1656(01)00407-2
Yang SJ, Lee HS, Park CS, Kim YR, Moon TW, Park KH (2004) Enzymatic analysis of an amylolytic enzyme from the hyperthermophilic archaeon Pyrococcus furiosus reveals its novel catalytic properties as both an α-amylase and a cyclodextrin-hydrolyzing enzyme. Appl Environ Microbiol 70:5988–5995. doi:10.1128/aem.70.10.5988-5995.2004
Acknowledgments
This work was financially supported by the Malaysian Ministry of Higher Education, Fundamental Research Grant Scheme (FRGS) and Universiti Teknologi Malaysia internal grant. We thank Dr. Stefan Janecek and Prof. Bernard Henrissat for their helpful discussion.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10295_2011_1074_MOESM1_ESM.pdf
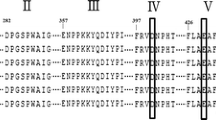
Amino acid sequence alignment between ASKA, ADTA, and known α-amylases. Enzyme sources are abbreviated as follows: ASKA, strain SK3-4; ADTA, strain DT3-1; BLA, Bacillus licheniformis (P06278); BSTA, Bacillus stearothermophilus (P06279); TAKA, Aspergillus oryzae (P0C1B3); and AmyA, Anaerobranca gottschalkii (Q5I942). The seven highly conserved regions among amylases are boxed; the three active site residues are highlighted; and the different amino acids between ASKA and ADTA are shown in white text (PDF 74 kb)
Rights and permissions
About this article
Cite this article
Chai, Y.Y., Rahman, R.N.Z.R.A., Illias, R.M. et al. Cloning and characterization of two new thermostable and alkalitolerant α-amylases from the Anoxybacillus species that produce high levels of maltose. J Ind Microbiol Biotechnol 39, 731–741 (2012). https://doi.org/10.1007/s10295-011-1074-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-1074-9