Abstract
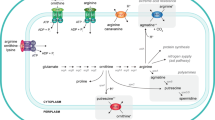
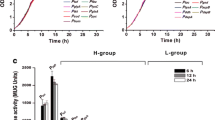
Corynebacterium glutamicum, a Gram-positive bacterium, has been widely used for industrial amino acid production. We previously showed that, in C. glutamicum, argCJBDFRGH arginine biosynthesis genes are clustered but independently transcribed from argC and argG promoters, leading to the generation of two transcripts corresponding to argCJBDFR and argGH. In this report, we show the effect of the C. glutamicum ArgR repressor on argC and argG promoters by overexpressing or disrupting the argR gene. Gel filtration assay results indicate that native ArgR is a hexamer of equal subunits with molecular mass of 110 kDa. Protein sequence analysis revealed the presence of an “SR” (Ser57-Arg58) motif for the DNA binding site at the N-terminal region and the “GTIAGDDTV” motif for arginine binding and its oligomerization at the C-terminal region. An argC or argG promoter–lacZ fusion reporter assay and argR mutational analysis showed that transcription of the argCJBDFR arginine biosynthesis genes is regulated from the argC promoter by ArgR in cooperation with l-arginine in C. glutamicum. This finding was supported by the gel mobility–shift assay showing direct binding of hexameric ArgR to the argC promoter in the presence of l-arginine. Unexpectedly, argGH transcription was not responsive to the level of ArgR repressor and/or arginine. In a further study, a C. glutamicum argR mutant was constructed by disrupting the chromosomal argR gene to manufacture an improved arginine-producing strain. Arginine productivity was increased in the C. glutamicum argR mutant strain under conditions of both limited and excessive arginine.








Similar content being viewed by others
References
Alén C, Sherratt DJ, Colloms SD (1997) Direct interaction of aminopeptidase A with recombination site DNA in Xer site-specific recombination. EMBO 16:5188–5197
Batt CA, Follettie MT, Shin HK, Yeh P, Sinskey AJ (1985) Genetic engineering of Coryneform bacteria. Trends Biotechnol 3:305–310
Charlier D, Roovers M, Vliet FV, Boyen A, Cunin R, Nakamura Y, Glandorff N, Piérard A (1992) Arginine Regulon of Escherichia coli K-12: a study of repressor-operator interactions and of in vitro binding affinities versus in Vitro repression. J Mol Biol 226:367–386
Chen SH, Merican AF, Sherratt DJ (1997) DNA binding of Escherichia coli arginine repressor mutants altered in oligomeric state. Mol Microbiol 24:1143–1153
Chun JY, Lee EJ, Lee HS, Cheon CI, Min KH, Lee MS (1998) Molecular cloning and analysis of the argC gene from Corynebacterium glutamicum. Biochem Mol Biol Int 46:437–447
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Lsborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squres S, Sulston JE, Taylor K, Whitehead S, Barrell BG (1988) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544
Cunin R, Glansdorff N, Pierard A, Stalon V (1986) Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 50:314–352
Dion M, Charlier D, Wang H, Gigot D, Savchenko A, Hallet JN, Glansdorff N, Sakanyan V (1997) The highly thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interactions. Mol Microbiol 25:385–398
Floriano B, Herrero A, Flores E (1994) Analysis of expression of the argC and argD genes in the Cyanobacterium Anabaena sp. strain PCC7120. J Bacteriol 176:6397–6401
Gardan R, Rappont G, Débarbouillé M (1997) Expression of the rocDEF operon involved in arginine catabolism in Bacillus sutilis. J Mol Biol 249:843–856
Grandori R, Lavoie TA, Pflumm M, Tian G, Niersbach H, Maas WK, Fairman R, Carey J (1995) The DNA-binding domain of the hexameric arginine repressor. J Mol Biol 254:150–162
Haas D, Holloway BW (1977) The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet 154:7–22
Harris Z, Singer M (1998) Identification and characterization of the Myxococcus xanthus argE gene. J Bacteriol 180:6412–6414
Hodgman TC, Griffiths H, Summers DK (1998) Nucleoprotein architecture and ColE1 dimer resolution: a hypothesis. Mol Microbiol 29:545–558
Hui S, Haifeng W, Diliana D, Vehary S, Nicolas G, Daniel C (2002) Transcription regulation in thermophilic Bacteria: high resolution contact probing of Bacillus stearothermophilus and Thermotoga neapolitana arginine repressor-operator interactions. J Mol Biol 315:255–274
Ikeda M, Nakagawa S (2003) The Corynebacterium glutamicum genome; Features and impacts on biotechonolgical processes. Appl Microbiol Biotechnol 62:99–109
Itoh Y (1997) Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol 197:7280–7290
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Keilhauer C, Eggeling L, Sahm H (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603
Kinoshita S (1985) Glutamic acid bacteria. In: Demain AL, Solomon NA (eds) Biology of industrial microorganisms. Benjamin/Commings, London, pp 115–142
Lee HS, Sinskey AJ (1994) Molecular characterization of AceB, a gene encoding malate synthase in Corynebacterium glutamicum. J Microbiol Biotechnol 4:256–263
Liebl W, Klamer R, Schleifer KH (1989) Requirement of chelating compounds for the growth of Corynebacterium glutamicum in synthetic media. Appl Microbiol Biotechnol 32:205–210
Lim D, Oppenheim JD, Eckhardt T, Maas WK (1987) Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc Natl Acad Sci USA 84:6697–6701
Maghnouj A, De Sousa Cabral TF, Stalon V, Wauven CV (1998) The arcABCD Gene Cluster, encoding the Arginine Deminase Pathway of Bacillus licheniformis, and Its Activation by the Arginine Repressor ArgR. J Bacteriol 180:6468–6475
Martin JF (1989) Molecular genetics of amino acid-producing Corynebacteria. In: Baumberg S, Hunter I, Rhodes M (eds) H. I. R. M. e. S. f. G. M. S. 4. Cambridge University Press, Cambridge, pp 25–59
Miller CM, Baumberg S, Stockley PG (1997) Operator interactions by the Bacillus subtilis arginine repressor activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol Microbiol 26:37–48
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Nishijyo T, Park SM, Lu CD, Itoh Y, Abdelal AT (1998) Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J Bacteriol 180:5559–5566
Park SM, Lu CD, Abdelal AT (1997) Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol 179:5300–5308
Park SM, Lu CD, Abdelal AT (1997) Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol 179:5309–5317
Rodríguez-García A, Lndovice M, Martin JF, Liras P (1997) Arginine boxes and the argR gene in Streptomyces clavuligerus: evidence for a clear regulation of the arginine pathway. Mol Microbiol 25:219–228
Sakanyan V, Petrosyan P, Lecocq M, Boyen A, Legrain C, Demarez M, Hallet JN, Glansdorff M (1996) Genes and enzymes of the acetyl cycle of arginine biosynthesis in Corynebacteirum glutamicum: enzyme evolution in the early steps of the arginine pathway. Microbiology 142:99–108
Sambrook J, Russell D (2001) Molecular cloning. A laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Schafer A, Kalinowski J, Simon R, Seep-Feldhaus A-H, Puhler A (1990) High-frequency conjugal plasmid transfer from gram-negative Escherichia coli to various gram-positive coryneform bacteria. J Bacteriol 172:1663–1666
Schwarzer A, Puhler A (1991) Manipulation of Corynebacterium glutamicum by gene disruption and replacement. Biotechnology 2:84–87
Simons R, Houman F, Kleckner N (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penaltics and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tian G, Lim D, Carey J, Mass WK (1992) Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J Mol Biol 226:387–397
Tian G, Maas WK (1994) Mutational analysis of the arginine repressor of Escherichia coli. Mol Microbiology 13:599–608
Udaka S (1966) Pathway-specific pattern of control of arginine biosynthesis in bacteria. J Bacteriol 91:617–621
Van de Casteele M, Dmarez M, Legrain C, Glansdorff N, Pierard A (1990) Pathways of arginine biosynthesis in extreme thermophilic archaeo- and eubacteria. J Gen Microbiol 136:1177–1183
Xu Y, Liang Z, Legrain C, Ruger J, Glansdorff N (2000) Evolution of arginine biosynthesis in the bacterial domain: novel gene-enzyme relationships from psychrophilic Moritella strains (Vibrionaceae) and evolutionary significance of N-alpha-acetyl ornithinase. J Bacteriol 182:1609–1615
Yim SH, Kim SH, Park MY, Jung SI, Lee HS, Cheon CI, Song ES, Lee SS, Lee MS (2008) Analysis of gene expression in arginine biosynthetic gene cluster of C. glutamicum. Genes Genomics 30:261
Acknowledgments
This work was supported by Sookmyung Women’s University (2010).
Author information
Authors and Affiliations
Corresponding author
Additional information
S.-H. Yim and S. Jung contributed equally to this paper.
Rights and permissions
About this article
Cite this article
Yim, SH., Jung, S., Lee, Sk. et al. Purification and characterization of an arginine regulatory protein, ArgR, in Corynebacterium glutamicum . J Ind Microbiol Biotechnol 38, 1911–1920 (2011). https://doi.org/10.1007/s10295-011-0977-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-0977-9