Abstract
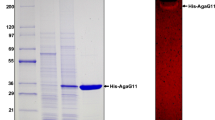
A gene (agrP) encoding a β-agarase from Pseudoalteromonas sp. AG4 was cloned and expressed in Escherichia coli. The agrP primary structure consists of an 870-bp open reading frame (ORF) encoding 290 amino acids (aa). The predicted molecular mass and isoelectric point were determined at 33 kDa and 5.9, respectively. The signal peptide was predicted to be 21 aa. The deduced aa sequence showed 98.6% identity to β-agarase from Pseudoalteromonas atlantica. The recombinant protein was purified as a fusion protein and biochemically characterized. The purified β-agarase (AgaP) had specific activity of 204.4 and 207.5 units/mg towards agar and agarose, respectively. The enzyme showed maximum activity at 55°C and pH 5.5. It was stable at pH 4.5 to 8.0 and below 55°C for 1 h. The enzyme produced neoagarohexaose and neoagarotetraose from agar and in addition to that neoagarobiose from the agarose. The neoagarooligosaccharides were biologically active. Hence, AgaP is a useful enzyme source for use by cosmetic and pharmaceutical industries.










Similar content being viewed by others
References
Craigie JS (1990) Cell walls. In: Cole KM, Sheath RG (eds) Biology of the red algae. Cambridge University Press, Cambridge, pp 221–257
Usov AI (1998) Structural analysis of red seaweed galactans of agar and carrageenan groups. Food Hydrocol 12:301–308
Araki C, Arai K (1967) Studies on the chemical constitution of agar-agar XXIV. Isolation of a new disaccharide as a reversion product from acidic hydrolysate. Bull Chem Societ Japan 40:1452–14564
Potin P, Richard C, Rochas C, Kloareg B (1993) Purification and characterization of the alpha-agarase from Alteromonas agarlyticus (Cataldi) comb. nov., strain GJ1B. Eur J Biochem 214:599–607
Araki C (1959) Seaweed polysaccharides. In: Wolfrom ML (ed) Carbohydrate chemistry of substances of biological interests. Pergamon Press, London, pp 15–306
Hassairi I, Ben AR, Nonus M, Gupta BB (2001) Production and separation of alpha-agarase from Alteromonas agarlyticus strain GJ1B. Biores Technol 79:47–517
Kobayashi R, Takisada M, Suzuki T, Kirimura K, Usami S (1997) Neoagarobiose as a novel moisturizer with whitening effect. Biosci Biotechnol Biochem 61:162–1638
Aoki T, Araki T, Kitamikado M (1990) Purification and characterization of a novel beta-agarase from Vibrio sp. AP-2. Eur J Biochem 187:461–465
Sugano Y, Terada I, Arita M, Noma M, Matsumoto T (1993) Purification and characterization of a new agarase from a marine bacterium, Vibrio sp. strain JT0107. Appl Environ Microbiol 59:1549–155410
Araki T, Lu Z, Morishita T (1998) Optimization of parameters for isolation of protoplasts from Gracilaria verrucosa (Rhodophyta). J Mar Biotechnol 6:193–197
Fu W, Han B, Duan D, Liu W, Wang C (2008) Purification and characterization of agarases from a marine bacterium Vibrio sp. F-6. J Ind Microbiol Biotechnol 35:915–922
Groleau D, Yaphe W (1977) Enzymatic hydrolysis of agar: purification and characterization of beta-neoagarotetraose hydrolase from Pseudomonas atlantica. Can J Microbiol 23:672–679
Morrice LM, McLean MW, Williamson FB, Long WF (1983) Beta-agarases I and II from Pseudomonas atlantica. Purifications and some properties. Eur J Biochem 135:553–558
Ha JC, Kim JT, Kim SK, Oh TK, Yu JH, Kong IS (1997) Beta agarase from Pseudomonas sp. W7: purification of the recombinant enzyme from Escherichia coli and the effects of salt on its activity. Biotechnol Appl Biochem 26:1–6
Vera J, Alvarez R, Murano E, Slebe JC, Leon O (1998) Identification of a marine agarolytic Pseudoalteromonas isolate and characterization of its extra cellular agarase. Appl Environ Microbiol 64:4378–4383
Schroeder DC, Jaffer MA, Coyne VE (2003) Investigation of the role of a beta (1–4) agarase produced by Pseudoalteromonas gracilis B9 in eliciting disease symptoms in the red alga Gracilaria gracilis. Microbiol 149:2919–2929
Leon O, Quintana L, Peruzzo G, Slebe JC (1992) Purification and properties of an extracellular agarase from Alteromonas sp. strain C-1. Appl Environ Microbiol 58:4060–4063
Kirimura K, Masuda N, Iwasaki Y, Nakagawa H, Kobayashi R, Usami S (1999) Purification and characterization of a novel beta agarase from an alkalophilic bacterium, Alteromonas sp. E-1. J Biosci Bioeng 87:436–441
Wang J, Mou H, Jiang X, Guan H (2006) Characterization of a novel beta-agarase from marine Alteromonas sp. SY37–12 and its degrading products. Appl Microbiol Biotechnol 71:833–839
Fu XT, Lin H, Kim SM (2008) Purification and characterization of a novel beta-agarase, AgaA34, from Agarivorans albus YKW-34. Appl Microbiol Biotechnol 78:265–273
Hu Z, Lin BK, Xu Y, Zhong MQ, Liu GM (2009) Production and purification of agarase from a marine agarolytic bacterium Agarivorans sp. HZ105. J Appl Microbiol 106:181–190
Duckworth M, Turvey JR (1969) The action of a bacterial agarase on agarose, porphyran and alkali treated porphyran. Biochem J 113:687–692
Van der Meulen HJ, Harder W (1975) Production and characterization of the agarase of Cytoplaga flevensis. Antonie van Leeuwenhoek 41:431–447
Ekborg NA, Taylor LE, Longmire AG, Henrissat B, Weiner RM, Hutcheson SW (2006) Genomic and proteomic analyses of the agarolytic system expressed by Saccharophagus degradans 2–40. Appl Environ Microbiol 72:3396–3405
Suzuki H, Sawai Y, Suzuki T, Kawai K (2003) Purification and characterization of an extracellular beta agarase from Bacillus sp. MK03. J Biosci Bioeng 93:456–463
Lee DG, Park GT, Kim NY, Lee EJ, Jang MK, Shin YG, Park GS, Kim TM, Lee JW, Lee JH, Kim SJ, Lee SH (2006) Cloning, expression, and characterization of a glycoside hydrolase family 50 beta-agarase from a marine Agarivorans isolate. Biotechnol Lett 28:1925–1932
Ohta Y, Hatada Y, Ito S, Horikoshi K (2005) High-level expression of a neoagarobiose-producing beta-agarase gene from Agarivorans sp. JAMB-A11 in Bacillus subtilis and enzymic properties of the recombinant enzyme. Biotechnol Appl Biochem 41:183–191
Ohta Y, Hatada Y, Nogi Y, Li Z, Ito S, Horikoshi K (2004) Cloning, expression, and characterization of a glycoside hydrolase family 86 beta agarase from a deep-sea Microbulbifer-like isolate. Appl Microbiol Biotechnol 66:266–275
Ohta Y, Hatada Y, Nogi Y, Miyazaki M, Li Z, Akita M, Hidaka Y, Goda S, Ito S, Horikoshi K (2004) Enzymatic properties and nucleotide and amino acid sequences of a thermostable beta-agarase from a novel species of deep-sea Microbulbifer. Appl Microbiol Biotechnol 64:505–514
Ohta Y, Hatada Y, Nogi Y, Miyazaki M, Li Z, Hatada Y, Ito S, Horikoshi K (2004) Enzymatic properties and nucleotide and amino acid sequences of a thermostable beta agarase from the novel marine isolate, JAMB-A94. Biosci Biotechnol Biochem 68(5):1073–1081
Ma C, Lu X, Shi C, Shi C, Li J, Gu Y, Ma Y, Chu Y, Han F, Gong Q (2007) Molecular cloning and characterization of a novel beta-agarase, AgaB, from marine Pseudoalteromonas sp. CY24. J Biol Chem 282:3747–3754
Lee S, Park J, Yoon S, Kim J, Kong I (2000) Sequence analysis of a beta-agarase gene (pjaA) from Pseudomonas sp. isolated from marine environment. J Biosci Bioeng 89:485–488
Ryu SK, Cho SJ, Park SR, Lim WJ, Kim MK, Hong SY, Baed W, Park YW, Kim BK, Kim H, Yun HD (2001) Cloning of the cel9A gene and characterization of its gene product from marine bacterium Pseudomonas sp. SK38. Appl Microbiol Biotechnol 57:138–145
Zhang WW, Sun L (2007) Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134. Appl Environ Microbiol 73:2825–2831
Kendall K, Cullum J (1984) Cloning and expression of an extracellular-agarase from Streptomyces coelicolor A3(2) in Streptomyces lividans 66. Gene 29:315–321
Jam M, Flament D, Allouch J, Potin P, Thion L, Kloareg B, Czjzek M, Helbert W, Michel G, Barbeyron T (2005) The endo-beta-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem J 385:703–713
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Pearson WR (1990) Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol 183:63–98
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic alignment search tool. J Mol Biol 215:403–410
Allouch J, Jam M, Helbert W, Barbeyron T, Kloareg B, Henrissat B, Czjzek M (2003) The three-dimensional structures of two beta agarases. J. Biol Chem 278:47171–47180
Michel G, Chantalat L, Duee E, Barbeyron T, Henrissat B, Kloareg B, Dideberg O (2001) The kappa-carrageenase of Pseudoalteromonas carrageenovora features a tunnel-shaped active site: a novel insight in the evolution of clan-B glycoside hydrolases. Structure 9:513–525
Allouch J, Helbert W, Henrissat B, Czjzek M (2004) Parallel substrate binding sites in a β-agarase suggest a novel mode of action on double-helical agarose. Structure 12:623–632
Keitel T, Meldgaard M, Heineman U (1994) Cation binding to a Bacillus (1, 3–1, 4)-β-glucanase Geometry, affinity and effect on protein stability. Eur J Biochem 222:203–214
Lee DG, Jang MK, Lee OH, Kim NY, Ju S, Lee SH (2008) Over-production of a glycoside hydrolase family 50 β-agarase from Agarivorans sp. JA-1 in Bacillus subtilis and the whitening effect of its product. Biotechnol Lett 30:911–918
Chen HM, Zheng L, Yan XJ (2005) The preparation and bioactivity research of agaro-oligosaccharides. Food Technol Biotechnol 43(1):29–36
Acknowledgment
This research was financially supported by the Ministry of Education, Science Technology (MEST) and Korea Institute for Advancement of Technology (KIAT) through the Human Resource Training Project for Regional Innovation, and by a research grant (PP00740) from Korea Ocean Research & Development Institute.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Oh, C., Nikapitiya, C., Lee, Y. et al. Cloning, purification and biochemical characterization of beta agarase from the marine bacterium Pseudoalteromonas sp. AG4. J Ind Microbiol Biotechnol 37, 483–494 (2010). https://doi.org/10.1007/s10295-010-0694-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0694-9