Abstract
A novel fibrinolytic enzyme (AJ) was purified from Staphylococcus sp. strain AJ screened from Korean salt-fermented Anchovy-jeot. Relative molecular weight of AJ was determined as 26 kDa by using SDS-PAGE and fibrin zymography. Based on a 2D gel, AJ was found to consist of three active isoforms (pI 5.5–6.0) with the same N-terminal amino acid sequence. AJ exhibited optimum pH and temperature at 2.5–3.0 and 85°C, respectively. AJ kept 85% of the initial activity after heating at 100°C for 20 min on the zymogram gel. The Michaelis constant (K m) and K cat values of AJ towards α-casein were 0.38 mM and 19.73 s−1, respectively. AJ cleaved the Aα-chain of fibrinogen but did not affect the Bβ- and γ-chains, indicating that it is an α-fibrinogenase. The fibrinolytic activity was inhibited by diisopropyl fluorophosphate, indicating AJ is a serine protease. Interestingly, AJ was very stable at acidic condition, SDS, and heat (100°C), whereas it was easily degraded at neutral and alkaline conditions. In particular, AJ formed an active homo-dimer in the pH range from 7.0 to 8.0. To our knowledge, a similar combination of acid and heat stability has not yet been reported for other fibrinolytic enzymes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fibrin is the primary protein component of a blood clot, which is formed from fibrinogen by the activated thrombin (EC 3.4.21.5). It is dissolved by fibrinolytic enzyme, plasmin (EC 3.4.21.7), which maintains blood flow at vascular injury sites and is an important component of the normal hemostatic response. When clots are not lysed, they accumulate in blood vessels, and cause thrombosis leading to myocardial infarction and other cardiovascular diseases [1, 2]. The major thrombolytic agents are classified into two types. The plasminogen activators, such as urokinase, tPA (tissue type plasminogen activator), and streptokinase, which activate plasminogen to plasmin, and the plasmin-like proteins, such as nattokinase [3] and lumbrokinase [4], which can directly degrade the fibrin.
Many bacterial fibrinolytic enzymes were discovered from fermented foods, such as natto [3, 5–7], shiokara [8] in Japanese food, Chungkook-Jang [1], Doen-Jang [2], and Jeot-Gal [9] in Korean food, douchi [10] in Chinese food, and Tempeh [11] in Indonesian food. Of them Shiokara (Japan) and Jeot-Gal (Korea) are fish-fermented foods, which are used as important additive for improving the taste of other foods as well as being foods themselves [12]. In addition, some fibrinolytic enzymes were discovered from root tissue of Stemona japonica (Blume) Miq, a Chinese traditional medicine [13], and marine alga (e.g., Codium latum, C. divaricatum, and C. intricatum [14–16]. In particular, regarding nattokinase produced by Bacillus natto screened from natto it was reported that the enzyme not only hydrolyzed thrombi in vivo, but also converted plasminogen to plasmin [6]. Oral administration of the enzyme showed that it could enhance fibrinolytic activity in plasma and the production of tPA, and its fibrinolytic activity was retained in the blood for more than 3 h.
Anchovy-jeot is a typical and popular fish-fermented food and is considered safe for long-term consumption. Recently, we isolated a halotolerant Staphylococcus strain, which produce an extracellular fibrinolytic enzyme, and designated as Staphylococcus sp. strain AJ. Here we report the purification, some characteristics and primary structure of the fibrinolytic enzyme (AJ) produced by Staphylococcus sp. strain AJ.
Materials and methods
Materials
Human fibrinogen, thrombin, plasmin, and diisopropyl fluorophosphates were purchased from Sigma (St Louis, MO, USA). Polyvinylidene difluoride (PVDF) membrane and electrophoretic reagents were purchased from Bio-Rad (Hercules, CA, USA). Other chemicals were of analytical grade.
Bacterial strain and cultural conditions
The bacterium isolated from Anchovy-jeot, which was confirmed by the Korean Collection for Type Cultures (KCTC) as a strain of Staphylococcus sp. strain AJ was grown in tryptic soy broth (TSB, Difco, Sparks, MD, USA) at 37°C for 2 days.
Purification of the fibrinolytic enzyme
The fibrinolytic enzyme (AJ) was purified from the culture supernatants of Staphylococcus sp. strain AJ. All purification procedures were performed at 4°C and with 20 mM sodium phosphate buffer, pH 6.4 (buffer A). Two liters of the crude enzyme was concentrated by ultrafiltration with the PM-10 membrane (Amicon Inc., USA). The concentrated solution was dialyzed against 20 volumes of buffer A for 1 day with three buffer changes. The dialyzed solution was loaded onto the DEAE-cellulose column (2.0 × 10 cm) (Phamacia Biotech, Sweden), and the proteins were fractionated with 300 ml linear gradient from 0 to 1 M NaCl. Fractions showing the fibrinolytic activity were pooled and dialyzed against buffer A, and then were concentrated by lyophilization. The fibrinolytic enzyme was further purified by the TSK gel filtration (Toyopearl HW-55F, TOSOH, Japan) using the buffer A containing 0.1 M NaCl.
Fibrin/casein plates
Fibrinolytic activity was determined by using fibrin plate method [17]. Fibrinogen (5 ml of 0.6% [w/v]) solution in a 50 mM sodium phosphate buffer (pH 7.4) was mixed with the same volume of 2% (w/v) agarose solution and 0.1 ml of thrombin (10 NIH U/ml) in a Petri dish. The solution was left for 1 h at room temperature to form a fibrin clot layer. Caseinolytic activity was assayed by using the casein plate. Casein (5 ml of 0.6% [w/v]) solution in a 50-mM sodium phosphate buffer (pH 7.4) was mixed with the same volume of 2% (w/v) agarose solution in a Petri dish. Twenty microliters (3 μg) of the sample solution was then dropped into holes that had been made in the fibrin plate by a capillary glass tube (5-mm diameter), and the plate was incubated at 37°C for 12 h. An equal volume of plasmin solution (1 NIH U/ml) also was incubated as a control. Enzyme activity was estimated by measuring the width of the clear zone. Each value represents the mean of four determinations.
Enzyme activity
Enzyme activity was assayed by the following procedure. A mixture (1.0 ml) containing 0.7 ml of 50 mM glycine–NaOH buffer (pH 10.0), 0.1 ml of 2% α-casein (w/v), and 0.1 ml of enzyme solution was incubated at 37°C for 10 min, mixed with 0.1 ml of 1.5 M trichloroacetic acid, allowed to stand at 37°C for 30 min, and then centrifuged at room temperature. The absorbance at 275 nm for the supernatant was measured and converted to the amount of tyrosine equivalent. The kinetic constants (K m) for the hydrolysis by the AJ were determined by Lineweaver–Burk plot using the various concentrations (1–10 mg/ml) of α-casein. One unit of caseinolytic activity was defined as the amount of enzyme releasing 1 μmol of tyrosine equivalent per minute.
Effect of pH on activity
The pH effect on the enzyme was assayed from pH 2.0 to 12.0. The enzyme in 20 mM sodium phosphate buffer (pH 6.4) was mixed with different buffers. The buffers used (and their pH ranges) were 0.1 M citrate–phosphate buffer (pH 2.0–5.0), sodium phosphate buffer (pH 6.0–7.0), Tris–HCl buffer (pH 8.0–9.0), and glycine–NaOH buffer (pH 10.0–12.0). The reaction mixtures were incubated at 37°C for 1 h, and the enzyme activities were measured using α-casein. Maximum activity was expressed as 100%, and the others were compared to the maximum activity. Each value represents the mean of four determinations.
Effect of temperature on activity
The effect of temperature on activity was assayed at various temperatures at pH 3.0. The enzyme in 20 mM citrate–phosphate buffer (pH 3.0) was incubated at different temperatures varying from 25 to 100°C for 20 min. The enzyme activities were assayed using α-casein, and the relative activities were calculated as percentage of the maximum activity.
Effects of protease inhibitors and metal ions
Each inhibitor and various divalent ions were mixed with purified enzyme (in 20 mM citrate–phosphate buffer, pH 3.0) at 37°C for 10 min. The enzyme activities were assayed using α-casein, and the relative activities were calculated on the basis of the activity of the pure enzyme without any inhibitor or metal ions under the same experimental conditions.
Zymography
Fibrin zymogram gel electrophoresis was carried out as described previously [18]. The separating gel solution (12%, w/v) was prepared in the presence of fibrinogen (0.12%, w/v) and 100 μl of thrombin (10 NIH U/ml). After electrophoresis in a cold room (at 10 mA constantly), the gel was incubated for 30 min in 50 mM Tris (pH 7.4), which contained 2.5% Triton ×-100. The gel was washed with distilled water for 30 min to remove Triton ×-100, and then incubated in zymogram reaction buffer (30 mM Tris, pH 7.4, and 0.02% [w/v] NaN3) at 37°C for 12 h. The gel was stained with Coomassie blue for 1 h and then destained. The bands indicating areas where fibrin was digested were visualized as the non-stained regions of the zymogram gel.
Two-dimensional gel electrophoresis
Isoelectic focusing (IEF) was carried out using 7-cm linear immobiline IPG gels with pH range of 3–10 (Bio-Rad, Hercules, CA, USA). Purified protein was loaded by in-gel rehydration with a reswelling solution containing 8 M urea, 0.3% DTT (w/v), and 0.2% (v/v) pH 3–10 IPG buffer. IEF was carried out for 4,000 Vh at 20°C in a IPGphor IEF system (Bio-Rad), wherein the voltage was linearly increased from 250 to 4,000 V over the first 3 h and then was maintained at 4,000 V for the final 10 h. After the IEF, the strips were equilibrated with the SDS equilibration buffer (62.5 mM Tris–HCl, pH 6.8, 2.3% [w/v] SDS, 30% [v/v] glycerol, and 0.05% [w/v] bromophenol blue) for 15 min. The equilibrated strips were loaded into the stacking gel of the SDS gel and fibrin gel, and the 2D gel electrophoresis was performed.
Fibrinogenolytic activity
Fibrinogenolytic activity [19] was assayed by incubating 0.1 ml of a 0.2% human fibrinogen solution (w/v) with 0.05 ml of enzyme solution (100 ng) in 50 mM Tris–HCl buffer (pH 7.4) at 37°C. At different times (0–5 min), 0.15 ml of a denaturing SDS sample buffer (0.125 mM Tris–HCl, pH 6.8, 0.1% [w/v] SDS, and 1% [w/v] β-mercaptoethanol) was added and the mixture heated at 95°C for 4 min. Twenty-five microliters (about 15 μg of fibrinogen) of each sample was subsequently analyzed by SDS-PAGE.
Determination of N-terminal amino acid sequence of AJ
After SDS-PAGE was done, purified protein on the gel was transferred to a PVDF membrane by electroblotting [20] and stained with Coomassie blue. The stained portion was excised and used for N-terminal sequencing directly by the automated Edman degradation method using a gas-phase protein sequencer (model Procise 491, ABI, USA).
PCR cloning of AJ gene
Using the amino acid sequence of AJ, we searched the nucleotide sequence database of National Center for Biotechnology Information for a AJ homologue and found the glutamyl endopeptidase clone (Accession No.: AB096695.1). Chromosomal DNA from Staphylococcus sp. strain AJ was prepared by the method of Rochelle [21], and used as the template for PCR. AJ gene was amplified by PCR using Nde I-linked sense primer (5′-GGAATTCCATATGGTAATATTACCTAATAATAATAGAC-3′) and Xho I-linked antisense primer (5′-CCGCTCGAGTTACTGAATATTTATATCAGGTATA-3′). PCR amplification was performed under the following conditions: 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min. The PCR-amplified 651 bp DNA fragment was extracted from agarose gel and then ligated into pGEM-T Easy vector (Promega) to generate pT-AJ plasmid.
Thrombolytic activity of the purified enzyme
For the thrombolytic test by purified enzyme, the actual fibrin clots (thrombi) of whole blood isolated from a rat’s vena cava were used [22]. Thrombi were prepared by adding of thrombin (10 NIH U/ml) to rat whole blood and then washing three times with 0.1 M Tris–HCl (pH 7.4) containing 0.9% (w/v) NaCl.
Results
Purification of the AJ from Staphylococcus sp. strain AJ
A bacterial strain producing extracellular fibrinolytic enzyme was isolated from Anchovy-jeot, a popular fish-fermented food in Korea. AJ was purified from the culture supernatants of Staphylococcus sp. strain AJ. Its purification procedure is summarized in Table 1. The final specific activity of the purified enzyme increased by more than 4.9-fold. AJ migrated as a single band, and its apparent molecular mass was estimated to be approximately 26 kDa by SDS and zymogram gel (Fig. 1a, b). Using a 2D gel, AJ was found to consist of three active isoforms (pI 5.5–6.0) with the same N-terminal amino acid sequence (Fig. 1c, d). Plus, AJ migrated as a single band on a native gel, which indicates they are homogenous protein (Fig. 2).
Analysis of AJ protein on a native gel (6%). After incubation of AJ at pH 3.0 or 7.0 for 2 h, the samples were diluted with five times sample buffer (without SDS and reducing agent). (312.5 mM Tris–HCl, pH 6.8, 50% glycerol, and 0.05% bromophenol blue). 1, 2 mean AJ at pH 3 and 7, respectively. M markers from the Sigma nondenatured protein molecular weight marker
Enzymatic characterization of AJ
The effect of pH on the activity of purified enzyme was examined. The enzyme was active in the broad pH range (Fig. 3a), and the optimum pH and temperature for the fibrinolytic activity were at 2.5–3.0 and 85°C, respectively (Fig. 3a, b). The thermal stability of the enzyme was examined by assaying the residual enzyme activity after the incubation of the enzyme at the various temperatures in 0.1 M citrate–phosphate buffer (pH 3.0). After incubation for 20 h at 25°C, the enzyme remained relatively stable (Fig. 3c), and also after incubation for 60 min at 70–80°C, the enzyme was fairly stable (Fig. 3d). Finally, the effect of protease inhibitors was studied at pH 3.0. The activity of the enzyme was inhibited by 1 mM DFP, but not affected by PMSF, EDTA, EGTA, E-64, or leupeptin (Table 2). In view of the effects of pH, temperature, and inhibitors, the enzyme can apparently be classified as a thermoacid-stable serine protease. Enzyme activity was also assayed in the presence of different metal ions at a concentration of 5 mM. Most of the metal ions studied had no significant effect on the enzyme, with the exception of Fe2+ with which the activity was about twofold lower (Table 2).
Effects of pH (a) and temperature (b) on enzyme activity of AJ from Staphylococcus sp. strain AJ. Effect of temperature on the activity and stability of AJ at pH 3.0. After incubation for 20-h at 25°C, the residual activity was measured (c). To establish the thermostability of AJ (d), the residual enzyme activity was measured after incubation for 60 min. Filled circle 70, open circle 80, filled inverted triangle 90, open inverted triangle 100°C
Stability of AJ on the zymogram and SDS gels
To study the primary structure of AJ under various pH conditions, SDS gel and fibrin zymography were performed. AJ kept 85% of the initial activity after heating at 100°C for 20 min on the zymogram gel (Fig. 4a). In particular, AJ was very stable at acidic condition, whereas it was easily degraded at neutral and alkaline conditions (Fig. 4b). Interestingly, in the pH range from 7.0 to 8.0, AJ formed a dimer, showing an activity on SDS gel and fibrin zymogram gel (Fig. 4b, c). However, when AJ was incubated with SDS solution (1–5%) or at 100°C for 20 min at pH 7.0, it was not degraded and showed the same pattern with acid condition (Figs. 5, 6).
Stability of AJ on the pH and temperature by using fibrin zymography (I) and SDS-PAGE (II). a After incubation of AJ at the indicated temperature for 20 min, the samples were applied on the 12% gels. 1 Molecular mass marker; 2–9 30, 40, 50, 60, 70, 80, 90, 100°C. b After incubation of AJ at the various pH for 1 h. 1 Molecular mass marker, 2–10 pH 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0. c Stability of AJ at pH 7.0. 1 Molecular mass marker; 2–7 after incubation of AJ at pH 7.0 for 0, 10, 20, 30, 40, 60 min. The positions of amino acid sequence were determined based on the AJ protein (deposited in GenBank under accession number AY829230)
Comparison of AJ with other proteases for fibrinolytic activity
The fibrinolytic activity of AJ was compared with those of other proteases. AJ was 1.2- and 1.6-fold less efficient than those of subtilisin Carlsberg and subtilisin BPN’, respectively (Table 3). On the other hand, the specific activity (F/C, the ratio of fibrinolytic to caseinolytic activity) was 2.8 and 1.6 times higher than those of subtilisin Carlsberg and subtilisin BPN’, respectively (Table 3). The K m and K cat values of AJ were 0.38 mM and 19.73 s−1, respectively (Table 4).
Fibrinogenolytic activity of AJ
AJ cleaved the Aα-chain of fibrinogen, but did not affect the Bβ- and γ-chains, indicating that AJ is a α-fibrinogenase (Fig. 7a). Even after a prolonged incubation of 10 min, no fibrinogen-clotting activity was observed (Fig. 7b).
SDS-polyacrylamide gel electrophoresis of the digested human fibrinogen by AJ under reducing conditions. a Fibrinogen consisted of three-polypeptide chains: Aα (66,000), Bβ (54,000), and γ (48,000). Fifteen microliters of fibrinogen incubated with enzymes were subjected to electrophoresis on 10% gel: 1 molecular mass marker; 2 fibrinogen incubated without enzyme for 5 min; 3–8 fibrinogen samples after incubation with enzymes for 0, 1, 2, 3, 4, 5 min. b After incubation of AJ with fibrinogen for 10 min, 100 μl of thrombin (1.0 NIH U/ml) was added. 1 Without AJ and 2 AJ (1.0 μg)
N-terminal amino acid sequence of the purified enzyme
The N-terminal amino acid sequence of the purified enzyme up to residue 17 was Val-Ile-Leu-Pro-Asn-Asn-X-Arg-His-Gln-Ile-Phe-X-Thr-Thr-Gln-Gly-, and shared a high degree of similarity with the N-terminal sequences of a glutamyl endopeptidase (also known as V8 serine protease) from S. epidermidis ATCC 12228 [23], S. aureus ATCC 12600 [24], and S. warneri M [25].
Cloning of AJ
AJ gene of Staphylococcus sp. strain AJ was cloned and its nucleotide sequence was determined (deposited in GenBank under accession number AY829230). The nucleotide sequence revealed only one large open reading frame. The amino acid sequence of AJ was compared with the published sequences of the other proteins, and AJ was found to be identical to V8 protease from S. epidermidis ATCC 12228 [23], except for one amino acid substitution Ser44Asn (Fig. 8, arrow marked).
Comparison of the amino acid sequence of AJ with those of other proteases. AJ, Staphylococcus sp. strain AJ (this study); V8, from S. epidermidis ATCC 12228 [23]; glutamic, glutamic acid-specific endoprotease from S. aureus ATCC 12600 [24]; glutamyl, glutamyl endopeptidase from S. warneri M [25]. Underline indicates the N-terminal amino acid position of 11 kDa fragment of AJ
Thrombolytic activity of the purified enzyme
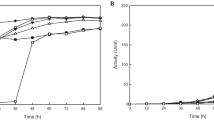
For the thrombolytic test by purified enzyme, the actual fibrin clots (thrombi) of whole blood isolated from a rat’s vena cava were used. As shown in Fig. 9b, the purified enzyme dissolved actual fibrin clots, indicating that AJ is a potent thrombolytic agent.
Discussion
This article describes the purification, some characteristics and primary structure of AJ produced from Staphylococcus sp. strain AJ for assessment of its application as a thrombosis agent. On the basis of this report, bacterial strain producing fibrinolytic enzyme was isolated from Anchovy-joet, a popular fish-fermented food in Korea, and was identified as Staphylococcus sp. strain AJ. AJ was purified from supernatant of Staphylococcus sp. strain AJ culture broth and it showed thermoacid-stable and strong fibrinolytic activity. But, no effect on plasminogen activator activity was observed.
In the pH range from 7.0 to 8.0, AJ formed an active homo-dimer on SDS gel and fibrin zymogram gel (Fig. 4b, c). We found that AJ consists of three isoforms (pI 5.5–6.0) with the same molecular mass and N-terminal amino acid sequence (Fig. 1c, d). Therefore, it is supposed that these three isoforms interact and form a dimer of AJ protein. Also, AJ was easily degraded as two fragments, 15 and 11 kDa, at neutral and alkaline conditions. However, no activity was found in both fragments. The N-terminal amino acid sequences of two fragments were determined (GenBank under accession number AY829230).
AJ showed unusual thermostable at acid condition, as shown in Figs. 3d and 4a, whereas it was unstable at neutral and alkaline conditions (Fig. 4b, c). We also examined the stability of AJ in SDS (1–5%) or incubation at 100°C for 20 min. Interestingly, AJ was not degraded and showed the same pattern with acid condition (Figs. 5, 6). This means that SDS and heating may act as a stabilizing factor. To our knowledge, a similar combination of acid and heat stability has not yet been reported for other fibrinolytic enzymes.
As mentioned above, AJ cleaved the Aα-chain of fibrinogen but did not affect the Bβ- and γ-chains (Fig. 7). In general, fibrin (ogen)olytic enzymes belong to as two classes, the α(β)-fibrinogenases (known as zinc-metalloproteinases) and the β-fibrinogenases (known as thermostable serine proteinases) [26, 27]. On the basis of these results, AJ is a new type of serine α-fibrinogenase.
In general, S. aureus secretes four major extracellular proteases: serine protease (V8 protease; SspA), cysteine protease (SspB), metalloprotease (aureolysin; Aur), and second cysteine protease (Scp). All four proteases are produced as proenzymes, which are proteolytically cleaved to form the active mature enzymes [28]. V8 serine protease of them was one of the first isolated enzymes of S. aureus to be purified and characterized in detail [29]. Its molecular mass was estimated to be 11.4 kDa by SDS gel, and it exhibited maximum proteolytic activity at pH 4.0 and 7.8. In recent, the glutamyl endopeptidase (designated GluSE), which showed 78.9% (15 residues) homology to that of mature GluV8, was purified from S. epidermidis ATCC 14990 [30]. GluSE was a 27 kDa and the optimum pH for activity was 8.0, and was low when under pH 6.0 or over pH 9.0, showing quite different enzymatic properties with AJ.
AJ gene of Staphylococcus sp. strain AJ was cloned and its nucleotide sequence was determined. The nucleotide sequence revealed only one large open reading frame. The deduced amino acid sequence of AJ was 99.5 similar to that of V8 protease from S. epidermidis ATCC 12228 (Fig. 8) [23]. But, two proteins showed different optimum pH condition, thermostability and substrate specificity. Clearly, change in amino acid at position 44 (Ser44Asn) may account for these differences. It is reported that the amino acid substitutions of Asp44Glu, Asn71Ser, and Lys147Arg of V8 protease increased the half-life of the enzyme stability against 5 M urea [31, 32]. Thus, site-directed mutagenesis and functional analysis of AJ will be continued to identify the amino acid critical for the thermoacid stability and substrate specificity.
References
Kim W, Choi K, Kim Y, Park H, Choi J, Lee Y, Oh H, Kwon I, Lee S (1996) Purification and characterization of a fibrinolytic enzyme produced from Bacillus sp. Strain CK 11-4 screened from Chungkook-Jang. Appl Environ Microbiol 62:2482–2488
Kim SH, Choi NS (2000) Purification and characterization of subtilisin DJ-4 secreted by Bacillus sp. strain DJ-4 screened from Doen-Jang. Biosci Biotechnol Biochem 64:1722–1725. doi:10.1271/bbb.64.1722
Sumi H, Hamada H, Tsushima H, Mihara H, Muraki H (1987) A novel fibrinolytic enzyme (nattokinase) in the vegetable cheese natto; a typical and popular soybean food in the Japanese diet. Experientia 43:1110–1111. doi:10.1007/BF01956052
Mihara H, Sumi H, Yoneta T, Mizumoto H, Ikeda R, Seiki M, Maruyama M (1991) A novel fibrinolytic enzyme extracted from the earthworm, Lumbricus rubellus. Jpn J Physiol 41:461–472. doi:10.2170/jjphysiol.41.461
Sumi H, Hamada H, Nakanishi K, Hiratani H (1990) Enhancement of the fibrinolytic activity in plasma by oral administration of nattokinase. Acta Haematol 84:139–143
Fujita M, Nomura K, Hong K, Ito Y, Asada A, Nishimuro S (1993) Purification and characterization of a strong fibrinolytic enzyme (nattokinase) in the vegetable cheese natto, a popular soybean fermented food in Japan. Biochem Biophys Res Commun 197:1340–1347. doi:10.1006/bbrc.1993.2624
Chang CT, Fan MH, Kuo FC, Sung HY (2000) Potent fibrinolytic enzyme from a mutant of Bacillus subtilis IMR-NK1. J Agric Food Chem 48:3210–3216. doi:10.1021/jf000020k
Sumi H, Nakajima N, Yatagai C (1995) A unique strong fibrinolytic enzyme (Katsuwokinase) in Skipjack “shiokara”, a Japanese traditional fermented food. Comp Biochem Physiol Biochem Mol Biol 112:543–547. doi:10.1016/0305-0491(95)00100-X
Kim HK, Kim GT, Kim DK, Choi WA, Park SH, Jeong YK, Kong IS (1997) Purification and characterization of a novel fibrinolytic enzyme from Bacillus sp. KA38 originated from fermented fish. J Ferment Bioeng 84:307–312. doi:10.1016/S0922-338X(97)89249-5
Peng Y, Huang Q, Zhang R, Zhang Y (2003) Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douche, a traditional Chinese soybean food. Comp Biochem Physiol Biochem Mol Biol 134:45–52. doi:10.1016/S1096-4959(02)00183-5
Kim SB, Lee DW, Cheigh CI, Choe EA, Lee SJ, Hong YH, Choi HJ, Pyun YR (2006) Purification and characterization of a fibrinolytic subtilisin-like protease of Bacillus subtilis TP-6 from an Indonesian fermented soybean, Tempeh. J Ind Microbiol Biotechnol 33:436–444. doi:10.1007/s10295-006-0085-4
Yoon JH, Kang SS, Lee KC, Kho YH, Choi SH, Kang KH, Park YH (2001) Bacillus jeotgali sp. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douche, a traditional Chinese soybean food. xInt J Syst Evol Microbiol 51:1087–1092
Lu F, Sun L, Lu Z, Bie X, Fang Y, Liu S (2007) Isolation and identification of an endophytic strain EJS-3 producing novel fibrinolytic enzyme. Curr Microbiol 54:435–439. doi:10.1007/s00284-006-0591-7
Matsubara K, Sumi H, Hori K, Miyazawa K (1998) Purification and characterization of two fibrinolytic enzymes from a marine green alga, Codium intricatum. Comp Biochem Physiol Biochem Mol Biol 119:177–181. doi:10.1016/S0305-0491(97)00303-9
Matsubara K, Hori K, Matsuura Y, Miyazawa K (1999) A fibrinolytic enzyme from a marine green alga, Codium latum. Phytochemistry 52:993–999. doi:10.1016/S0031-9422(99)00356-8
Matsubara K, Hori K, Matsuura Y, Miyazawa K (2000) Purification and characterization of a fibrinolytic enzyme and identification of fibrinogen clotting enzyme in a marine green alga, Codium divaricatum. Comp Biochem Physiol Biochem Mol Biol 125:137–143. doi:10.1016/S0305-0491(99)00161-3
Asrup T, Müllertz S (1952) The fibrin plate method for estimating fibrinolytic activity. Arch Biochem Biophys 40:346–351. doi:10.1016/0003-9861(52)90121-5
Kim SH, Choi NS, Lee WY (1998) Fibrin zymography: a direct analysis of fibrinolytic enzyme on gels. Anal Biochem 263:115–116. doi:10.1006/abio.1998.2816
Hung CC, Huang KF, Chiou SH (1994) Characterization of one novel venom protease with β-fibrinogenase activity from the Taiwan habu (Trimeresurus mucrosquamatus): purification and cDNA sequence analysis. Biochem Biophys Res Commun 205:1707–1715. doi:10.1006/bbrc.1994.2865
Matsudaira P (1987) Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membrane. J Biol Chem 262:10035–10038
Rochelle PA, Fry JC, Parkes RJ, Weightman AJ (1992) DNA extraction for 16S rRNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett 100:59–66. doi:10.1111/j.1574-6968.1992.tb05682.x
Nakajima N, Ishihara K, Sugimoto M, Sumi H, Mikuni K, Hamata H (1996) Chemical modification of earthworm fibrinolytic enzyme with human serum albumin fragment and characterization of the protease as a therapeutic enzyme. Biosci Biotechnol Biochem 60:293–300
Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM (2003) Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49:1577–1593. doi:10.1046/j.1365-2958.2003.03671.x
Yoshikawa K, Tsuzuki H, Fujiwara T, Nakamura E, Iwamoto H, Matsumoto K, Shin M, Yoshida N, Teraoka H (1992) Purification, characterization and gene cloning of a novel glutamic acid-specific endopeptidase from Staphylococcus aureus ATCC 12600. Biochim Biophys Acta 1121:221–228
Yokoi K, Kakikawa M, Kimoto H, Watanabe K, Yasukawa H, Yamakawa A, Taketo A, Kodaira I (2001) Genetic and biochemical characterization of glutamyl endopeptdase of Staphylococcus warneri M. Gene 281:115–122. doi:10.1016/S0378-1119(01)00782-X
Siigur E, Aaspõllu A, Tu AT, Siigur J (1996) cDNA cloning and deduced amino acid sequence of fibrinolytic enzyme (lebetase) from Vipera lebetina snake venom. Biochem Biophys Res Commun 224:229–236. doi:10.1006/bbrc.1996.1012
Lee JW, Seu JH, Rhee IK, Jin I (1999) Purification and characterization of brevinase, a heterogeneous two-chain fibrinolytic enzyme from the venom of Korean snake, Agkistrodon blomhoffii brevicaudus. Biochem Biophys Res Commun 260:665–670. doi:10.1006/bbrc.1999.0977
Karlsson A, Arvidson S (2002) Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect Immun 70:4239–4246. doi:10.1128/IAI.70.8.4239-4246.2002
Drapeau GR, Boily Y, Houmard J (1972) Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem 247:6720–6726
Ohara-Nemoto Y, Ikeda Y, Kobayashi M, Sasaki M, Tajika S, Kimura S (2002) Characterization and molecular cloning of a glutamyl endopeptidase from Staphylococcus epidermidis. Microb Pathog 33:33–41. doi:10.1006/mpat.2002.0515
Yabuta M, Onai-Miura S, Ohsuye K (1995) Isolation and characterization of urea-resistant Staphylococcus aureus V8 protease derivatives. J Ferment Bioeng 80:237–243. doi:10.1016/0922-338X(95)90822-H
Yabuta M, Ohsuye K (1995) Increase in urea-resistance of recombinant V8 protease by combining mutations, and its application in the releasing of a peptide hormone from a susion protein. J Ferment Bioeng 80:467–472. doi:10.1016/0922-338X(96)80921-4
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Choi, NS., Song, J.J., Chung, DM. et al. Purification and characterization of a novel thermoacid-stable fibrinolytic enzyme from Staphylococcus sp. strain AJ isolated from Korean salt-fermented Anchovy-joet . J Ind Microbiol Biotechnol 36, 417–426 (2009). https://doi.org/10.1007/s10295-008-0512-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0512-9