Abstract
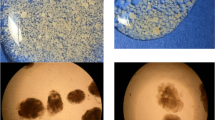
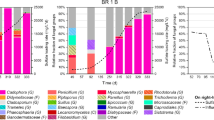
An anaerobic down-flow fluidized bed reactor was inoculated with granular sludge and started-up with sulfate containing synthetic wastewater to promote the formation of a biofilm enriched in sulfate-reducing bacteria (SRB), to produce biogenic sulfide. The start-up was done in two stages operating the reactor in batch for 45 days followed by 85 days of continuous operation. Low-density polyethylene was used as support. The biofilm formation was followed up by biochemical and electron microscopy analyses and the composition of the community was examined by 16S rDNA sequence analysis. Maximum immobilized volatile solids (1.2 g IVS/Lsupport) were obtained after 14 days in batch regime. During the 85 days of continuous operation, the reactor removed up to 80% of chemical oxygen demand (COD), up to 28% of the supplied sulfate and acetate was present in the effluent. Sulfate-reducing activity determined in the biofilm with ethanol or lactate as substrate was 11.7 and 15.3 g COD/g IVS per day, respectively. These results suggested the immobilization of sulfate reducers that incompletely oxidize the substrate to acetate; the phylogenetic analysis of the cloned 16S rDNA gene sequences showed high identity to the genus Desulfovibrio that oxidizes the substrates incompletely. In contrast, in the granular sludge used as inoculum a considerable number of clones showed homology to Methanobacterium and just few clones were close to SRB. The starting-up approach allowed the enrichment of SRB within the diverse community developed over the polyethylene support.





Similar content being viewed by others
References
Acuña N, Ortega-Morales BO, Valadez-González A (2006) Biofilm colonization dynamics and its influence on the corrosion resistance of austenitic UNSS31603 stainless steel exposed to Gulf of Mexico seawater. Mar Biotechnol 8:62–70. doi:10.1007/s10126-005-5145-7
Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Alvarez MT, Pozzo T, Mattiasson B (2006) Enhancement of sulphate production in anaerobic packed bed bench-scale biofilm reactors by sulphate reducing bacteria. Biotechnol Lett 28:175–181. doi:10.1007/s10529-005-5332-7
Alves CF, Melo LF, Vieira MJ (2002) Influence of medium composition on the characteristics of a denitrifying biofilm formed by Alcaligenes denitrificans in a fluidised bed reactor. Process Biochem 37:837–845. doi:10.1016/S0032-9592(01)00282-5
Amann RI, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
APHA (1998) Standard methods for the examination of water and wastewater. American Public Health Association, Washington DC
Barns SM, Fundyga RE, Jeffries MW, Pace NR (1994) Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA 91:1609–1613. doi:10.1073/pnas.91.5.1609
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. doi:10.1093/nar/7.6.1513
Bond PL, Smriga SP, Bandfield JF (2000) Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl Environ Microbiol 66:3842–3849. doi:10.1128/AEM.66.9.3842-3849.2000
Boonapatcharoen N, Meepian K, Chaiprasert P, Techkarnjanaruk S (2007) Molecular monitoring of microbial population dynamics during operational periods of anaerobic hybrid reactor treating Cassava starch wastewater. Microb Ecol 54:21–30. doi:10.1007/s00248-006-9161-6
Briones AM, Daugherty BJ, Angement LT, Rausch KD, Tumbleson ME, Raskin L (2007) Microbial diversity and dynamics in multi-and single-compartment anaerobic bioreactors processing sulfate-rich waste streams. Environ Microbiol 9:93–106. doi:10.1111/j.1462-2920.2006.01119.x
Buffière P, Bergeon JP, Moletta R (2000) The inverse turbulent bed: a novel bioreactor for anaerobic treatment. Water Res 34:673–677. doi:10.1016/S0043-1354(99)00166-9
Bryers JD (2000) Process engineering. In: Bryers JD (ed) Biofilms II: process analysis and applications. Wiley-Liss, New York, pp 13–44
Celis-García LB, González-Blanco G, Meraz M (2008) Removal of sulfur inorganic compounds by a biofilm of sulfate reducing and sulfide oxidizing bacteria in a down-flow fluidized bed reactor. J Chem Technol Biotechnol 83:260–268. doi:10.1002/jctb.1802
Cord-Ruwisch R (1985) A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J Microbiol Methods 4:33–36. doi:10.1016/0167-7012(85)90005-3
Devereux R, Stahl DA (1993) Phylogeny of sulfate-reducing bacteria and a perspective for analyzing their natural communities. In: Odom JM, Singleton RJ (eds) The sulfate-reducing bacteria: contemporary perspectives. Springer, New York, pp 131–160
Flemming HC, Wingender J, Moritz R, Brochard W, Mayer C (1999) Physico-chemical properties of biofilms—a short review. In: Keevil CW, Godfree A, Holt D, Dow C (eds) Biofilms in the aquatic environment. Royal Society of Chemistry, London, pp 1–12
Freeman SA, Sierra-Alvarez R, Altinbas M, Hollingsworth J, Stams AJM, Smidt H (2008) Molecular characterization of mesophilic and thermophilic sulfate reducing microbial communities in expanded granular sludge bed (EGSB) reactors. Biodegradation 19:161–177. doi:10.1007/s10532-007-9123-9
Kaksonen AH, Riekkola-Vanhanen ML, Puhakka JA (2003) Optimization of metal sulphide precipitation in fluidized-bed treatment of acidic wastewater. Water Res 37:255–266. doi:10.1016/S0043-1354(02)00267-1
Kaksonen AH, Plumb JJ, Franzmann PD, Puhakka JA (2004) Simple organic electron donors support diverse sulfate-reducing communities in fluidized-bed reactors treating acidic metal- and sulfate-containing wastewater. FEMS Microbiol Ecol 47:279–289. doi:10.1016/S0168-6496(03)00284-8
Kwok WK, Picioreanu C, Ong SL, van Loosdrecht MCM, Ng WJ, Heijnen JJ (1998) Influence of biomass production and detachment forces on biofilm structures in a biofilm airlift suspension reactor. Biotechnol Bioeng 58:400–407. doi:10.1002/(SICI)1097-0290(19980520)58:4<400::AID-BIT7>3.0.CO;2-N
Lazarova V, Pierzo V, Fontvielle D, Manem J (1994) Integrated approach for biofilm characterisation and biomass activity control. Water Sci Technol 29(7):345–354
Lefebvre O, Vasudevan N, Thanasekaran K, Moletta R, Godon JJ (2006) Microbial diversity in hypersaline wastewater: the example of tanneries. Extremophiles 10:505–513. doi:10.1007/s00792-006-0524-1
Lens PNL, Van Den Bosch MC, Hulshoff Pol LW, Lettinga G (1998) Effect of staging on volatile fatty acid degradation in a sulfidogenic granular sludge reactor. Water Res 32:1178–1192. doi:10.1016/S0043-1354(97)00323-0
Liu H, Huang L, Huang Z, Zheng J (2007) Specification of sulfate reducing bacteria biofilms accumulation effects on corrosion initiation. Mater Corros 58:44–48. doi:10.1002/maco.200603984
Meraz M, Monroy O, Noyola A, Ilangovan K (1995) Studies on the dynamics of immobilization of anaerobic bacteria on a plastic support. Water Sci Technol 32(8):243–250. doi:10.1016/0273-1223(96)00032-7
Nagpal S, Chuichulcherm S, Peeva L, Livingston A (2000) Microbial sulfate reduction in a liquid-solid fluidized bed reactor. Biotechnol Bioeng 70:370–380. doi:10.1002/1097-0290(20001120)70:4<370::AID-BIT2>3.0.CO;2-7
Ohashi A, Harada H (1994) Adhesion strength of biofilm developed in an attached-growth reactor. Water Sci Technol 29(10–11):281–288
Oude Elferink SJWH, Vorstman WJC, Sopjes A, Stams AJM (1998) Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol Ecol 27:185–194. doi:10.1111/j.1574-6941.1998.tb00536.x
Silva AJ, Hirasawa JS, Varesche MB, Foresti E, Zaiat M (2006) Evaluation of support materials for the immobilization of sulfate-reducing bacteria and methanogenic archaea. Anaerobe 12:93–98. doi:10.1016/j.anaerobe.2005.12.003
van Benthum WAJ, Garrido-Fernández JM, Tijhuis L, van Loosdrecht MCM, Heijnen JJ (1996) Formation and detachment of biofilms and granules in a nitrifying biofilm airlift suspension reactor. Biotechnol Prog 12:764–772. doi:10.1021/bp960063e
Visser A, Beeksma I, van der Zee F, Stams AJM, Lettinga G (1993) Anaerobic degradation of volatile fatty acids at different sulphate concentrations. Appl Microbiol Biotechnol 40:549–556
Widdel F (1988) Microbiology and ecology of sulfate- and sulfur-reducing bacteria. In: Zehnder AJB (ed) Biology of anaerobic organisms. Wiley, New York, pp 469–586
Wisotzkey JD, Jurtshuk PJ, Fox GE (1990) PCR amplification of 16S rDNA from lyophilized cell cultures facilitates studies in molecular systematics. Curr Microbiol 21:325–327. doi:10.1007/BF02092099
Xu Q, Wen X, Deng X (2004) A simple protocol for isolating genomic DNA from chestnut rose (Rosa roxburghii Tratt) for RFP and PCR analyses. Plant Mol Biol Rep 22:301a–301g. doi:10.1007/BF02773140
Zehnder AJB, Huser BA, Brock TD, Wuhrmann K (1980) Characterization of an acetate-decarboxylating, non-hydrogen oxidizing methane bacterium. Arch Microbiol 124:1–11. doi:10.1007/BF00407022
Acknowledgments
This research was financially supported by the projects SEP-CONACyT-46506 and 62028. The authors acknowledge the technical assistance of Rosalba Castillo-Collazo, Dulce Partida-Gutierrez and Marisol Gallegos-Garcia. We also thank Dr. Mónica Meraz from the Department of Biotechnology at the Universidad Autónoma Metropolitana-Iztapalapa for her kind help with the design and construction of the DFFB reactor.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Celis, L.B., Villa-Gómez, D., Alpuche-Solís, A.G. et al. Characterization of sulfate-reducing bacteria dominated surface communities during start-up of a down-flow fluidized bed reactor. J Ind Microbiol Biotechnol 36, 111–121 (2009). https://doi.org/10.1007/s10295-008-0478-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0478-7