Abstract
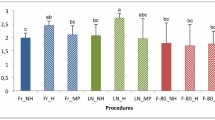
The widespread use of molecular techniques in studying microbial communities has greatly enhanced our understanding of microbial diversity and function in the natural environment and contributed to an explosion of novel commercially viable enzymes. One of the most promising environments for detecting novel processes, enzymes, and microbial diversity is hot springs. We examined potential biases introduced by DNA preservation and extraction methods by comparing the quality, quantity, and diversity of environmental DNA samples preserved and extracted by commonly used methods. We included samples from sites representing the spectrum of environmental conditions that are found in Yellowstone National Park thermal features. Samples preserved in a non-toxic sucrose lysis buffer (SLB), along with a variation of a standard DNA extraction method using CTAB resulted in higher quality and quantity DNA than the other preservation and extraction methods tested here. Richness determined using DGGE revealed that there was some variation within replicates of a sample, but no statistical difference among the methods. However, the sucrose lysis buffer preserved samples extracted by the CTAB method were 15–43% more diverse than the other treatments.



Similar content being viewed by others
References
Allen ET, Day AL (1935) Hot springs of the Yellowstone National Park. Carnegie Institution, Washington, DC
Barns SM, et al. (1994) Remarkable archaeal diversity in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA 91(March):1609–1613 doi:10.1073/pnas.91.5.1609
Braid MD, et al. Testing the UltraClean soil DNA purification kit on a diverse range of soils by PCR amplification of 16S rDNA. In: ASM General Meeting. 1999
Brock TD Thermophilic microorganisms and life at high temperatures. 1978, Springer-Verlag, New York, pp 465
Bryant DA et al (2007) Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523–526. doi:10.1126/science.1143236
Cary SC et al (1993) Identification and localization of bacterial endosymbionts in hydrothermal vent taxa with symbiont-specific polymerase chain reaction amplification and in situ hybridization techniques. Mol Mar Biol Biotechnol 2(1):51–62
Ferris MJ, Ward DM (1997) Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol 63(4):1375–1381
Frantzen MAJ et al (1998) Empirical evaluation of preservation methods for faecal DNA. Mol Ecol 7:1423–1428. doi:10.1046/j.1365-294x.1998.00449.x
Fromin N et al (2002) Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ Microbiol 4(11):634–643. doi:10.1046/j.1462-2920.2002.00358.x
Frostegard A et al (1999) Quantification of bias related to the extraction of DNA directly from soils. Appl Environ Microbiol 65(12):5409–5420
Gabor EM, de Vries EJ, Janssen DB (2003) Effecient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol Ecol 44:153–163. doi:10.1016/S0168-6496(02)00462-2
Garrett RH, Grisham CM (1995) In: Field C (ed) Biochemistry, Saunders College Publishing, Philadelphia
Ghosh D et al (2003) Molecular phylogenetic exploration of bacterial diversity in a Bakreshwar (India) hot spring and culture of Shewanella-related thermophiles. Appl Environ Microbiol 69(7):4332–4336. doi:10.1128/AEM.69.7.4332-4336.2003
Giovannoni SJ et al (1990) Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol 56:2572–2575
Haldeman DL et al (1994) Changes in bacteria recoverable from subsurface volcanic rock samples during storage at 4°C. Appl Environ Microbiol 60(8):2697–2703
Harris KJ, Kelley ST, Pace NR (2004) New perspective on uncultured bacterial phylogenetic division OP11. Appl Environ Microbiol 70(2):845–849. doi:10.1128/AEM.70.2.845-849.2004
Harry M, Gambier B, Garnier-Sillam E (2000) Soil conservation for DNA preservation for bacterial molecular studies. Eur J Soil Sci 36:51–55
Herrera A, Cockell CS (2007) Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbiol Methods 70:1–12. doi:10.1016/j.mimet.2007.04.005
Hobel CFV et al (2004) Use of low nutrient enrichments to access novel amylase genes in silent diversity of thermophiles. World J Microbiol Biotechnol 20:801–809. doi:10.1007/s11274-004-2623-4
Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180(18):4765–4774
Khanna M, Stotzky G (1992) Transformation of Bacillus subtilis by DNA bound on montmorillonite and effect of DNase on the transforming ability of bound DNA. Appl Environ Microbiol 58(6):1930–1939
Kowalchuk GA et al (2007) Finding the needles in the metagenome haystack. Microb Ecol 53:475–485. doi:10.1007/s00248-006-9201-2
LaMontagne MG et al (2002) Evaluation of extraction and purification methods for obtaining PCR-amplifiable DNA from compost for microbial community analysis. J Microbiol Methods 49:255–264. doi:10.1016/S0167-7012(01)00377-3
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Leuko S et al (2008) Lysis efficiency of standard DNA extraction methods for Halococcus spp. in an organic rich environment. Extremophiles 12:301–308. doi:10.1007/s00792-007-0124-8
Lorenz P, Eck J (2005) Metagenomics and industrial applications. Nat Rev Microbiol 3:510–516. doi:10.1038/nrmicro1161
Luna GM, Dell’Anno A, Danovaro R (2006) DNA extraction procedure: a critical issue for bacterial diversity assessment in marine sediments. Environ Microbiol 8(2):308–320. doi:10.1111/j.1462-2920.2005.00896.x
Mandrioli M, Borsatti F, Mola L (2006) Factors affecting DNA preservation from museum-collected lepidopteran specimens. Entomol Exp Appl 120(3):239–244. doi:10.1111/j.1570-7458.2006.00451.x
Meyer-Dombard DR, Shock EL, Amend JP (2005) Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology 3:211–227. doi:10.1111/j.1472-4669.2005.00052.x
Miller DN et al (1999) Evaluation and optimization of DNA extraction and purification procedures for soil and sediment samples. Appl Environ Microbiol 65(11):4715–4724
Moeseneder MM et al (1999) Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol 65(8):3518–3525
More MI et al (1994) Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol 60(5):1572–1580
Nakagawa S et al (2005) Sulfurihydrogenibium yellowstonense sp. nov., an extremely thermophilic, facultatively heterotrophic, sulfur-oxidizing bacterium from Yellowstone National Park, and emended descriptions of the genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int J Syst Evol Microbiol 55:2263–2268. doi:10.1099/ijs.0.63708-0
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740. doi:10.1126/science.276.5313.734
Pontes DS et al (2007) Molecular approaches: advantages and artifacts in assessing bacterial diversity. J Ind Microbiol Biotechnol 34:463–473. doi:10.1007/s10295-007-0219-3
Reysenbach AL, Ehringer M, Hershberger K (2000) Microbial diversity at 83 degrees C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4(1):61–67
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a Laboratory manual. 2nd edn, Plainview, Cold Spring Harbor Laboratory Press, New York
Schoenborn L et al (2004) Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl Environ Microbiol 70(7):4363–4366. doi:10.1128/AEM.70.7.4363-4366.2004
Schoenfeld T, et al. (2008) Viral diversity and improved DNA polymerases, In: YNP RCN/TBI workshop, Mammoth Hot Springs, WY, USA
Speksnijder AGCL et al (2001) Microvariation artifacts introduced by pcr and cloning of closely related 16S rRNA gene sequences. Appl Environ Microbiol 67(1):469–472. doi:10.1128/AEM.67.1.469-472.2001
Steffan RJ, Atlas RM (1998) DNA amplification to enhance detection of genetically engineered bacteria in environmental samples. Appl Environ Microbiol 54(9):2185–2191
Stein JL et al (1996) Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol 178(3):591–599
Takacs-Vesbach CD et al (2008) Volcanic calderas delineate biogeographic provinces among Yellowstone thermophiles. Environ Microbiol 10(7):1681–1689. doi:10.1111/j.1462-2920.2008.01584.x
von Wintzingerode F, Gobel UB, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21(3):213–229. doi:10.1111/j.1574-6976.1997.tb00351.x
Yang ZH et al (2007) Comparison of methods for total community DNA extraction and purification from compost. Appl Microbiol Biotechnol 74:918–925. doi:10.1007/s00253-006-0704-z
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62(2):316–322
Acknowledgments
We are grateful for the comments and input by Robert Sinsabaugh and Diana Northup who kindly reviewed an earlier version of the manuscript. This work was supported by NSF Biodiversity Surveys and Inventories grant 02-06773 to CTV.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mitchell, K.R., Takacs-Vesbach, C.D. A comparison of methods for total community DNA preservation and extraction from various thermal environments. J Ind Microbiol Biotechnol 35, 1139–1147 (2008). https://doi.org/10.1007/s10295-008-0393-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0393-y