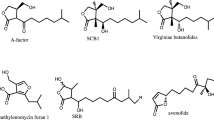
Abstract
Streptomyces clavuligerus produces a large array of natural compounds with antibiotic, antitumor, β-lactamase inhibition or inmunomodulating activities. The production of cephamycin C, clavulanic acid and other compounds with a clavam structure has been studied for many years. A network of regulatory mechanisms is present in S. clavuligerus to control the formation of different compounds by pathway-specific regulators or pleiotropic regulators. The possible existence of a γ-butyrolactone signaling system in this streptomycete is emerging. In addition, S. clavuligerus possesses a stringent control mechanism somehow different from those previously reported in other Streptomyces species.





Similar content being viewed by others
References
Alexander DC, Jensen SE (1998) Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J Bacteriol 180:4068–4079
Arias P, Fernández-Moreno MA, Malpartida F (1999) Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol 181:6958–6968
Arulanantham H, Kershaw NJ, Hewitson KS, Hughes CE, Thirkettle JE, Schofield CJ (2006) ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. J Biol Chem 6:279–287
Baggaley KH, Brown AG, Schofield CJ (1997) Chemistry and biosynthesis of clavulanic acid and other clavams. Natl Prod Rep 140:309–333
Bascarán V, Sánchez L, Hardisson C, Braña AF (1991) Stringent response and initiation of secondary metabolism in Streptomyces clavuligerus. J Gen Microbiol 137:1625–1634
Beck CF, Warren RA (1988) Divergent promoters, a common form of gene organization. Microbiol Rev 52:318–326
Belcourt MF, Farabaugh PJ (1990) Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 62:339–352
Bignell DR, Warawa JL, Strap JL, Chater KF, Leskiw BK (2000) Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology 146:2161–2173
Bignell DRD, Tahlan K, Colvin KR, Jensen SE, Leskiw BK (2005) Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on bldG. Antimicrob Agents Chemother 49:1529–1541
Brune I, Jochmann N, Brinkrolf K, Hüser AT, Gerstmeir R, Eikmanns BJ, Kalinowski J, Pühler A, Tauch A (2007) The IclR-type transcriptional repressor LtbR regulates the expression of leucine and tryptophan biosynthesis genes in the amino acid producer Corynebacterium glutamicum. J Bacteriol 189:2720–2733
Cashel M, Gentry DR, Hernandez VJ, Vinella D (1996) The Stringent Response. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington DC, pp 1458–1496
Champness WC (1988) New loci required for Streptomyces coelicolor morphological and physiological differentiation. J Bacteriol 170:1168–1174
Diederich B, Wilkinson JF, Magnin T, Najafi M, Erringston J, Yudkin MD (1994) Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor sigma F of Bacillus subtilis. Genes Dev 8:2653–2663
Duncan L, Losick R (1993) SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc Natl Acad Sci 90:2325–2329
Folcher M, Gaillard H, Nguyen LT, Nguyen KT, Lacroix P, Bamas-Jacques N, Rinkel M, Thompson CJ (2001) Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J Biol Chem 276:44297–44306
Fuente A de la, Lorenzana LM, Martín JF, Liras P (2002) Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J Bacteriol 184:6559–6565
Gomez-Escribano JP, Liras P, Pisabarro A, Martín JF (2006) An rplKΔ29−PALG−32 mutation leads to reduced expression of the regulatory genes ccaR and claR and very low transcription of the ceaS2 gene for clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol 61:758–770
Gomez-Escribano JP, Martin JF, Hesketh A, Bibb MJ, Liras P (2008) Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154:744–755
Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, Smith CP, Chater KF (2007) New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8:261
Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M (2007) The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol 8:R161
Hesketh A, Sun J, Bibb M (2001) Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol Microbiol 39:136–144
Horinouchi S (2002) A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front Biosci 7:2045–2057
Horinouchi S (2003) AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 30:462–467
Horinouchi S (2007) Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci Biotechnol Biochem 71:283–299
Ishida K, Hung TV, Liou K, Lee HC, Shin CH, Sohng JK (2006) Characterization of pbpA and pbp2 encoding penicillin-binding proteins located on the downstream of clavulanic acid gene cluster in Streptomyces clavuligerus. Biotechnol Lett 28:409–417
Jensen SE, Elder KJ, Aidoo KA, Paradkar AS (2000) Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob Agents Chemother 44:720–726
Jensen SE, Paradkar AS, Mosher RH, Anders C, Beatty PH, Brumlik MJ, Griffin A, Barton B (2004) Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother 48:192–202
Jin W, Ryu YG, Kang SG, Kim SK, Saito N, Ochi K, Lee SH, Lee KJ (2004) Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology 150:1485–1493
Jones D, Thompson A, England R (1996) Guanosine 5′-diphosphate 3′-diphosphate (ppGpp), guanosine 5′diphosphate 3′monophosphate (ppGp) and antibiotic production in Streptomyces clavuligerus. Microbiology 142:1789–1795
Kawamoto S, Zhang D, Ochi K (1998) Sequence analysis of the ribosomal L11 protein gene (rplK = relC) in Streptomyces lavendulae using a deletion allele. J Antibiot 51:954–957
Kenig M, Reading C (1979) Holomycin and an antibiotic (MM 19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J Antibiot 32:549–554
Kim D-W, Chater K, Lee K-J, Hesketh A (2005) Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J Bacteriol 187:2957–2966
Kim HS, Lee YJ, Lee CK, Choi SU, Yeo S, Hwang YI, Yu TS, Kinoshita H, Nihira T (2004) Cloning and characterization of a gene encoding the gamma-butyrolactone autoregulator receptor from Streptomyces clavuligerus. Arch Microbiol 182:44–50
Kim HS, Park YI (2007) Lipase activity and tacrolimus production in Streptomyces clavuligerus CKD1119 mutant strains. J Microbiol Biotechnol 17:1638–1644
Kondo K, Higuchi Y, Sakuda S, Nihira T, Yamada Y (1989) New virginiae butanolides from Streptomyces virginiae. J Antibiot 42:1873–1876
Kovacevic S, Tobin MB, Miller JR (1990) The beta-lactam biosynthesis genes for isopenicillin N epimerase and deacetoxycephalosporin C synthetase are expressed from a single transcript in Streptomyces clavuligerus. J Bacteriol 172:3952–3958
Kyung YS, Hu WS, Sherman DH (2001) Analysis of temporal and spatial expression of the CcaR regulatory element in the cephamycin C biosynthetic pathway using green fluorescent protein. Mol Microbiol 40:530–541
Lawlor E J, Baylis HA, Chater KF (1987) Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev 1:1305–1310
Li R, Khaleeli N, Townsend CA (2000) Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J Bacteriol 182:4087–4095
Liras P (1999) Biosynthesis and molecular genetics of cephamycins. Cephamycins produced by actinomycetes. Antonie Van Leeuwenhoek 75:109–124
Liras P, Rodríguez-García A (2000) Clavulanic acid, a beta-lactamase inhibitor: biosynthesis and molecular genetics. Appl Microbiol Biotechnol 54:467–475
Lorenzana LM, Pérez-Redondo R, Santamarta I, Martín JF, Liras P (2004) Two oligopeptide-permease-encoding genes in the clavulanic acid cluster of Streptomyces clavuligerus are essential for production of the beta-lactamase inhibitor. J Bacteriol 186:3431–3438
Mellado E, Lorenzana LM, Rodríguez-Sáiz M, Díez B, Liras P, Barredo JL (2002) The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 148:1427–1438
Molina-Henares AJ, Krell T, Guazzaroni ME, Segura A, Ramos JL (2006) Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev 30:157–186
Mori K (1983) Revision of the absolute configuration of A-factor, the inducer of streptomycin biosynthesis, basing on the reconfirmed (R)-configuration of (+)-paraconic acid. Tetrahedron 39:3107–3109
Mosher RH, Paradkar AS, Anders C, Barton B, Jensen SE (1999) Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob Agents Chemother 43:1215–1224
Ochi K (1986) Occurrence of the stringent response in Streptomyces sp. and its significance for the initiation of morphological and physiological differentiation. J Gen Microbiol 132:2621–2631
Ohnishi Y, Kameyama S, Onaka H, Horinouchi S (1999) The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol Microbiol 34:102–111
Ohnishi Y, Yamazaki H, Kato J, Tomono A, Horinouchi S (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439
Paradkar AS, Aidoo KA Jensen SE (1998) A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Mol Microbiol 27:831–843
Paradkar AS, Jensen SE (1995) Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J Bacteriol 177:1307–1314
Pérez-Llarena FJ, Liras P, Rodríguez-García A, Martín JF (1997) A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J Bacteriol 179:2053–2059
Pérez-Llarena FJ, Rodríguez-García A, Enguita FJ, Martín JF, Liras P (1998) The pcd gene encoding piperideine-6-carboxylate dehydrogenase involved in biosynthesis of alpha-aminoadipic acid is located in the cephamycin cluster of Streptomyces clavuligerus. J Bacteriol 180:4753–4756
Pérez-Redondo R, Rodríguez-García A, Martín JF, Liras P (1998) The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene 211:311–321
Recio E, Colinas A, Rumbero A, Aparicio JF, Martín JF (2004) PI factor, a novel type quorum-sensing inducer elicits pimaricin production in Streptomyces natalensis. J Biol Chem 279:41586–41593
Santamarta I, Rodríguez-García A, Pérez-Redondo R, Martín JF, Liras P (2002) CcaR is an autoregulatory protein that binds to the ccaR and cefD-cmcI promoters of the cephamycin C-clavulanic acid cluster in Streptomyces clavuligerus. J Bacteriol 184:3106–3113
Santamarta I, Pérez-Redondo R, Lorenzana LM, Martín JF, Liras P (2005) Different proteins bind to the butyrolactone receptor protein ARE sequence located upstream of the regulatory ccaR gene of Streptomyces clavuligerus. Mol Microbiol 56:824–835
Santamarta I, López-García MT, Pérez-Redondo R, Koekman B, Martín JF, Liras P (2007) Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the ARE ccaR sequence of S. clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Mol Microbiol 66:511–524
Sato K, Nihira T, Sakuda S, Yamagimoto M, Yamada Y (1989) Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Bioeng 68:170–173
Stutzman-Engwall KJ, Otten SL, Hutchinson CR (1992) Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol 174:144–154
Tahlan K, Anders C, Jensen SE (2004a) The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. J Bacteriol 186:6286–6297
Tahlan K, Park HU, Wong A, Beatty PH, Jensen SE (2004b) Two sets of paralogous genes encode the enzymes involved in the early stages of clavulanic acid and clavam metabolite biosynthesis in Streptomyces clavuligerus. Antimicrob Agents Chemother 48:930–939
Tahlan K, Anders C, Wong A, Mosher RH, Beatty PH, Brumlik MJ, Griffin A, Hughes C, Griffin J, Barton B, Jensen SE (2007) 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chem Biol 14:131–142
Takano E, Tao M, Long F, Bibb MJ, Wang L, Li W, Buttner MJ, Bibb MJ, Deng ZX, Chater KF (2003) A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol Microbiol 50:475–486
Takano E (2006) γ-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Curr Opin Microbiol 9:287–294
Tang L, Grimm A, Zhang YX, Hutchinson CR (1996) Purification and characterization of the DNA-binding protein DnrI, a transcriptional factor of daunorubicin biosynthesis in Streptomyces peucetius. Mol Microbiol 22:801–813
Trepanier NK, Jensen SE, Alexander DC, Leskiw BK (2002) The positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus is mistranslated in a bldA mutant. Microbiology 148:643–656
Wang L, Tahlan K, Kaziuk TL, Alexander DC, Jensen SE (2004) Transcriptional and translational analysis of the ccaR gene from Streptomyces clavuligerus. Microbiology 150:4137–4145
White J, Bibb M (1997) bldA dependence of undecylprodigiosin production in Streptomyces coelicolor A3(2) involves a pathway-specific regulatory cascade. J Bacteriol 179:627–633
Wietzorrek A, Bibb M (1997) A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol 25:1181–1184
Aknowledgments
This research was supported by grants from the CICYT (Madrid) (Proyecto Bio2006-14853) and by the European Proyect LSHM-CT-2004-005224. We thank Prof. A. L. Demain for correcting the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liras, P., Gomez-Escribano, J.P. & Santamarta, I. Regulatory mechanisms controlling antibiotic production in Streptomyces clavuligerus . J Ind Microbiol Biotechnol 35, 667–676 (2008). https://doi.org/10.1007/s10295-008-0351-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0351-8